Negative binomial regression is for modeling count variables, usually for over-dispersed count outcome variables.
This page uses the following packages. Make sure that you can load
them before trying to run the examples on this page. If you do not have
a package installed, run: install.packages("packagename"), or
if you see the version is out of date, run: update.packages().
require(foreign) require(ggplot2) require(MASS)
Version info: Code for this page was tested in R Under development (unstable) (2013-01-06 r61571)
On: 2013-01-22
With: MASS 7.3-22; ggplot2 0.9.3; foreign 0.8-52; knitr 1.0.5
Please note: The purpose of this page is to show how to use various data analysis commands. It does not cover all aspects of the research process which researchers are expected to do. In particular, it does not cover data cleaning and checking, verification of assumptions, model diagnostics or potential follow-up analyses.
Examples of negative binomial regression
Example 1. School administrators study the attendance behavior of high school juniors at two schools. Predictors of the number of days of absence include the type of program in which the student is enrolled and a standardized test in math.
Example 2. A health-related researcher is studying the number of hospital visits in past 12 months by senior citizens in a community based on the characteristics of the individuals and the types of health plans under which each one is covered.
Description of the data
Let’s pursue Example 1 from above.
We have attendance data on 314 high school juniors from two urban high schools in
the file nb_data. The response variable of interest is days absent, daysabs.
The variable math gives the standardized math score for
each student. The variable prog is a three-level nominal variable indicating the
type of instructional program in which the student is enrolled.
Let’s look at the data. It is always a good idea to start with descriptive statistics and plots.
dat <- read.dta("https://stats.idre.ucla.edu/stat/stata/dae/nb_data.dta") dat <- within(dat, { prog <- factor(prog, levels = 1:3, labels = c("General", "Academic", "Vocational")) id <- factor(id) }) summary(dat)
## id gender math daysabs ## 1001 : 1 female:160 Min. : 1.0 Min. : 0.00 ## 1002 : 1 male :154 1st Qu.:28.0 1st Qu.: 1.00 ## 1003 : 1 Median :48.0 Median : 4.00 ## 1004 : 1 Mean :48.3 Mean : 5.96 ## 1005 : 1 3rd Qu.:70.0 3rd Qu.: 8.00 ## 1006 : 1 Max. :99.0 Max. :35.00 ## (Other):308 ## prog ## General : 40 ## Academic :167 ## Vocational:107 ## ## ## ##
ggplot(dat, aes(daysabs, fill = prog)) + geom_histogram(binwidth = 1) + facet_grid(prog ~ ., margins = TRUE, scales = "free")
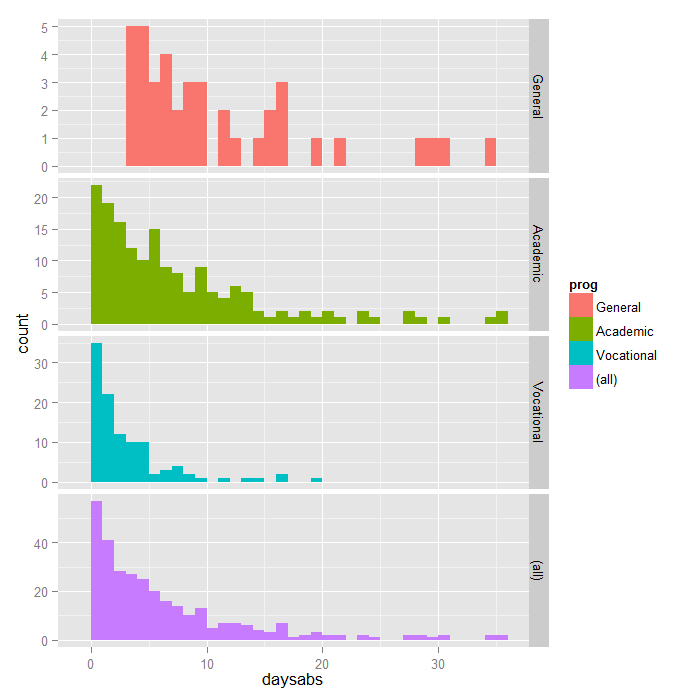
Each variable has 314 valid observations and their distributions seem quite reasonable. The unconditional mean of our outcome variable is much lower than its variance.
Let’s continue with our description of the variables in this dataset.
The table below shows the average numbers of days absent by program type
and seems to suggest that program type is a good candidate for predicting the number of
days absent, our outcome variable, because the mean value of the outcome appears to vary by
prog. The variances within each level of prog are
higher than the means within each level. These are the conditional means and
variances. These differences suggest that over-dispersion is present and that a
Negative Binomial model would be appropriate.
with(dat, tapply(daysabs, prog, function(x) { sprintf("M (SD) = %1.2f (%1.2f)", mean(x), sd(x)) }))
## General Academic Vocational ## "M (SD) = 10.65 (8.20)" "M (SD) = 6.93 (7.45)" "M (SD) = 2.67 (3.73)"
Analysis methods you might consider
Below is a list of some analysis methods you may have encountered. Some of the methods listed are quite reasonable, while others have either fallen out of favor or have limitations.
- Negative binomial regression -Negative binomial regression can be used for over-dispersed count data, that is when the conditional variance exceeds the conditional mean. It can be considered as a generalization of Poisson regression since it has the same mean structure as Poisson regression and it has an extra parameter to model the over-dispersion. If the conditional distribution of the outcome variable is over-dispersed, the confidence intervals for the Negative binomial regression are likely to be wider as compared to those from a Poisson regression model.
- Poisson regression – Poisson regression is often used for modeling count data. Poisson regression has a number of extensions useful for count models.
- Zero-inflated regression model – Zero-inflated models attempt to account for excess zeros. In other words, two kinds of zeros are thought to exist in the data, “true zeros” and “excess zeros”. Zero-inflated models estimate two equations simultaneously, one for the count model and one for the excess zeros.
- OLS regression – Count outcome variables are sometimes log-transformed and analyzed using OLS regression. Many issues arise with this approach, including loss of data due to undefined values generated by taking the log of zero (which is undefined), as well as the lack of capacity to model the dispersion.
Negative binomial regression analysis
Below we use the glm.nb function from the MASS package to
estimate a negative binomial regression.
summary(m1 <- glm.nb(daysabs ~ math + prog, data = dat))
## ## Call: ## glm.nb(formula = daysabs ~ math + prog, data = dat, init.theta = 1.032713156, ## link = log) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -2.155 -1.019 -0.369 0.229 2.527 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 2.61527 0.19746 13.24 < 2e-16 *** ## math -0.00599 0.00251 -2.39 0.017 * ## progAcademic -0.44076 0.18261 -2.41 0.016 * ## progVocational -1.27865 0.20072 -6.37 1.9e-10 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for Negative Binomial(1.033) family taken to be 1) ## ## Null deviance: 427.54 on 313 degrees of freedom ## Residual deviance: 358.52 on 310 degrees of freedom ## AIC: 1741 ## ## Number of Fisher Scoring iterations: 1 ## ## ## Theta: 1.033 ## Std. Err.: 0.106 ## ## 2 x log-likelihood: -1731.258
-
- R first displays the call and the deviance residuals. Next, we see the
regression coefficients for each of the variables, along with standard
errors, z-scores, and p-values. The variable
mathhas a coefficient of -0.006, which is statistically significant. This means that for each one-unit increase inmath, the expected log count of the number of days absent decreases by 0.006. The indicator variable shown asprogAcademicis the expected difference in log count between group 2 and the reference group (prog=1). The expected log count for level 2 ofprogis 0.44 lower than the expected log count for level 1. The indicator variable forprogVocationalis the expected difference in log count between group 3 and the reference group.The expected log count for level 3 ofprogis 1.28 lower than the expected log count for level 1. To determine ifprogitself, overall, is statistically significant, we can compare a model with and withoutprog. The reason it is important to fit separate models, is that unless we do, the overdispersion parameter is held constant.
- R first displays the call and the deviance residuals. Next, we see the
regression coefficients for each of the variables, along with standard
errors, z-scores, and p-values. The variable
m2 <- update(m1, . ~ . - prog) anova(m1, m2)
## Likelihood ratio tests of Negative Binomial Models ## ## Response: daysabs ## Model theta Resid. df 2 x log-lik. Test df LR stat. ## 1 math 0.8559 312 -1776 ## 2 math + prog 1.0327 310 -1731 1 vs 2 2 45.05 ## Pr(Chi) ## 1 ## 2 1.652e-10
- The two degree-of-freedom chi-square test indicates that
progis a statistically significant predictor ofdaysabs. - The null deviance is calculated from an intercept-only model with 313 degrees of freedom. Then we see the residual deviance, the deviance from the full model. We are also shown the AIC and 2*log likelihood.
- The theta parameter shown is the dispersion parameter. Note that R parameterizes this differently from SAS, Stata, and SPSS. The R parameter (theta) is equal to the inverse of the dispersion parameter (alpha) estimated in these other software packages. Thus, the theta value of 1.033 seen here is equivalent to the 0.968 value seen in the Stata Negative Binomial Data Analysis Example because 1/0.968 = 1.033.
Checking model assumption
As we mentioned earlier, negative binomial models assume the conditional means are not equal to the conditional variances. This inequality is captured by estimating a dispersion parameter (not shown in the output) that is held constant in a Poisson model. Thus, the Poisson model is actually nested in the negative binomial model. We can then use a likelihood ratio test to compare these two and test this model assumption. To do this, we will run our model as a Poisson.
m3 <- glm(daysabs ~ math + prog, family = "poisson", data = dat) pchisq(2 * (logLik(m1) - logLik(m3)), df = 1, lower.tail = FALSE)
## 'log Lik.' 2.157e-203 (df=5)
In this example the associated chi-squared value estimated from 2*(logLik(m1) – logLik(m3)) is 926.03 with one degree of freedom. This strongly suggests the negative binomial model, estimating the dispersion parameter, is more appropriate than the Poisson model.
We can get the confidence intervals for the coefficients by profiling the likelihood function.
(est <- cbind(Estimate = coef(m1), confint(m1)))
## Estimate 2.5 % 97.5 % ## (Intercept) 2.615265 2.2421 3.012936 ## math -0.005993 -0.0109 -0.001067 ## progAcademic -0.440760 -0.8101 -0.092643 ## progVocational -1.278651 -1.6835 -0.890078
We might be interested in looking at incident rate ratios rather than coefficients. To do this, we can exponentiate our model coefficients. The same applies to the confidence intervals.
exp(est)
## Estimate 2.5 % 97.5 % ## (Intercept) 13.6708 9.4127 20.3470 ## math 0.9940 0.9892 0.9989 ## progAcademic 0.6435 0.4448 0.9115 ## progVocational 0.2784 0.1857 0.4106
The output above indicates that the incident rate for prog = 2
is 0.64 times the incident rate for the reference group (prog = 1).
Likewise, the incident rate for prog = 3 is 0.28 times the incident
rate for the reference group holding the other variables constant. The
percent change in the incident rate of daysabs is a 1% decrease
for every unit increase in math.
The form of the model equation for negative binomial regression is the same as that for Poisson regression. The log of the expected outcome is predicted with a linear combination of the predictors:
\[ ln(\widehat{daysabs_i}) = Intercept + b_1I(prog_i = 2) + b_2I(prog_i = 3) + b_3math_i \]
where $I(prog_i = j)$ is an indicator function such that if $prog_i = j$ is equal to 1 and otherwise is equal to 0, for $j \in \{2, 3\}$. Therefore,
\[ \widehat{daysabs_i} = e^{Intercept + b_1 I(prog_i = 2) + b_2I(prog_i = 3) + b_3 math_i} = e^{Intercept}e^{b_1 I(prog_i = 2)}e^{b_2 I(prog_i = 3)}e^{b_3 math_i} \]
The coefficients have an additive effect in the $ln(y)$ scale
and the IRR have a multiplicative effect in the $y$ scale. The
dispersion parameter in negative binomial regression
does not affect the expected counts, but it does affect the estimated variance of
the expected counts. More details can be found in the Modern Applied
Statistics with S by W.N. Venables and B.D. Ripley (the book
companion of the MASS package).
For additional information on the various metrics in which the results can be presented, and the interpretation of such, please see Regression Models for Categorical Dependent Variables Using Stata, Second Edition by J. Scott Long and Jeremy Freese (2006).
Predicted values
For assistance in further understanding the model, we can look at predicted
counts for various levels of our predictors. Below we create new datasets with
values of math and prog and then use the predict command to
calculate the predicted number of events.
First, we can look at predicted counts for each value of prog while
holding math at its mean.
To do this, we create a new dataset with the combinations of prog and
math for which we would like to find predicted values, then use the predict
command.
newdata1 <- data.frame(math = mean(dat$math), prog = factor(1:3, levels = 1:3, labels = levels(dat$prog))) newdata1$phat <- predict(m1, newdata1, type = "response") newdata1
## math prog phat ## 1 48.27 General 10.237 ## 2 48.27 Academic 6.588 ## 3 48.27 Vocational 2.850
In the output above, we see that the predicted number of events (e.g., days
absent) for a general program is about 10.24, holding math at its mean. The predicted
number of events for an academic program is lower at 6.59, and the
predicted number of events for a vocational program is about 2.85.
Below we will obtain the mean predicted number of events for values of math
across its entire range for each level of prog and graph these.
newdata2 <- data.frame( math = rep(seq(from = min(dat$math), to = max(dat$math), length.out = 100), 3), prog = factor(rep(1:3, each = 100), levels = 1:3, labels = levels(dat$prog))) newdata2 <- cbind(newdata2, predict(m1, newdata2, type = "link", se.fit=TRUE)) newdata2 <- within(newdata2, { DaysAbsent <- exp(fit) LL <- exp(fit - 1.96 * se.fit) UL <- exp(fit + 1.96 * se.fit) }) ggplot(newdata2, aes(math, DaysAbsent)) + geom_ribbon(aes(ymin = LL, ymax = UL, fill = prog), alpha = .25) + geom_line(aes(colour = prog), size = 2) + labs(x = "Math Score", y = "Predicted Days Absent")
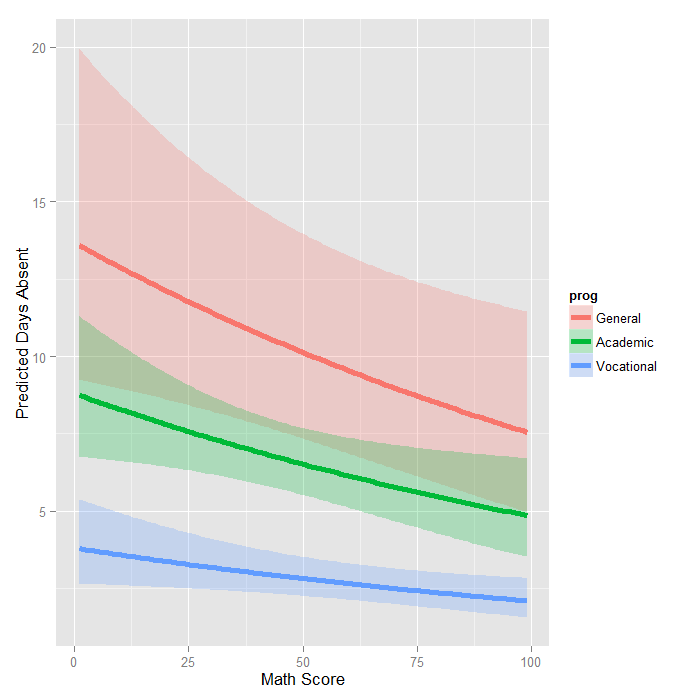
The graph shows the expected count across the range of math scores, for each type of program along with 95 percent confidence intervals. Note that the lines are not straight because this is a log linear model, and what is plotted are the expected values, not the log of the expected values.
Things to consider
- It is not recommended that negative binomial models be applied to small samples.
- One common cause of over-dispersion is excess zeros by an additional data generating process. In this situation, zero-inflated model should be considered.
- If the data generating process does not allow for any 0s (such as the number of days spent in the hospital), then a zero-truncated model may be more appropriate.
- Count data often have an exposure variable, which indicates the number
of times the event could have happened. This variable should be
incorporated into your negative binomial regression model with the use of
the
offsetoption. See theglmdocumentation for details. - The outcome variable in a negative binomial regression cannot have negative numbers.
- You will need to use the
m1$residcommand to obtain the residuals from our model to check other assumptions of the negative binomial model (see Cameron and Trivedi (1998) and Dupont (2002) for more information).
References
- Long, J. S. 1997. Regression Models for Categorical and Limited Dependent Variables. Thousand Oaks, CA: Sage Publications.
- Long, J. S. and Freese, J. 2006. Regression Models for Categorical Dependent Variables Using Stata, Second Edition. College Station, TX: Stata Press.
- Cameron, A. C. and Trivedi, P. K. 2009. Microeconometrics Using Stata. College Station, TX: Stata Press.
- Cameron, A. C. and Trivedi, P. K. 1998. Regression Analysis of Count Data. New York: Cambridge Press.
- Cameron, A. C. Advances in Count Data Regression Talk for the Applied Statistics Workshop, March 28, 2009. http://cameron.econ.ucdavis.edu/racd/count.html .
- Dupont, W. D. 2002. Statistical Modeling for Biomedical Researchers: A Simple Introduction to the Analysis of Complex Data. New York: Cambridge Press.
- Venables, W.N. and Ripley, B.D. 2002. Modern Applied Statistics with S, Fourth Edition. New York: Springer.
See also
- R online documentation: glm
