Probit regression, also called a probit model, is used to model dichotomous or binary outcome variables. In the probit model, the inverse standard normal distribution of the probability is modeled as a linear combination of the predictors.
This page uses the following packages. Make sure that you can load
them before trying to run the examples on this page. If you do not have
a package installed, run: install.packages("packagename"), or
if you see the version is out of date, run: update.packages().
require(aod) require(ggplot2)
Version info: Code for this page was tested in R Under development (unstable) (2012-11-16 r61126)
On: 2012-12-15
With: ggplot2 0.9.3; aod 1.3; knitr 0.9
Please Note: The purpose of this page is to show how to use various data analysis commands. It does not cover all aspects of the research process which researchers are expected to do. In particular, it does not cover data cleaning and checking, verification of assumptions, model diagnostics and potential follow-up analyses.
Examples
Example 1: Suppose that we are interested in the factors that influence whether a political candidate wins an election. The outcome (response) variable is binary (0/1); win or lose. The predictor variables of interest are the amount of money spent on the campaign, the amount of time spent campaigning negatively and whether the candidate is an incumbent.
Example 2: A researcher is interested in how variables, such as GRE (Graduate Record Exam scores), GPA (grade point average) and prestige of the undergraduate institution, effect admission into graduate school. The response variable, admit/don’t admit, is a binary variable.
Description of the Data
For our data analysis below, we are going to expand on Example 2 about getting into graduate school. We have generated hypothetical data, which can be obtained from our website in R. Note that R requires forward slashes (/) not back slashes () when specifying a file location even if the file is on your hard drive.
mydata <- read.csv("https://stats.idre.ucla.edu/stat/data/binary.csv") ## convert rank to a factor (categorical variable) mydata$rank <- factor(mydata$rank) ## view first few rows head(mydata)
## admit gre gpa rank ## 1 0 380 3.61 3 ## 2 1 660 3.67 3 ## 3 1 800 4.00 1 ## 4 1 640 3.19 4 ## 5 0 520 2.93 4 ## 6 1 760 3.00 2
This data set has a binary response (outcome, dependent) variable called admit.
There are three predictor variables: gre, gpa and rank. We will treat the
variables gre and gpa as continuous. The variable rank takes on the
values 1 through 4. Institutions with a rank of 1 have the highest prestige,
while those with a rank of 4 have the lowest.
summary(mydata)
## admit gre gpa rank ## Min. :0.000 Min. :220 Min. :2.26 1: 61 ## 1st Qu.:0.000 1st Qu.:520 1st Qu.:3.13 2:151 ## Median :0.000 Median :580 Median :3.40 3:121 ## Mean :0.318 Mean :588 Mean :3.39 4: 67 ## 3rd Qu.:1.000 3rd Qu.:660 3rd Qu.:3.67 ## Max. :1.000 Max. :800 Max. :4.00
xtabs(~rank + admit, data = mydata)
## admit ## rank 0 1 ## 1 28 33 ## 2 97 54 ## 3 93 28 ## 4 55 12
Analysis methods you might consider
Below is a list of some analysis methods you may have encountered. Some of the methods listed are quite reasonable while others have either fallen out of favor or have limitations.
- Probit regression, the focus of this page.
- Logistic regression. A logit model will produce results similar probit regression. The choice of probit versus logit depends largely on individual preferences.
- OLS regression. When used with a binary response variable, this model is known as a linear probability model and can be used as a way to describe conditional probabilities. However, the errors (i.e., residuals) from the linear probability model violate the homoskedasticity and normality of errors assumptions of OLS regression, resulting in invalid standard errors and hypothesis tests. For a more thorough discussion of these and other problems with the linear probability model, see Long (1997, p. 38-40).
- Two-group discriminant function analysis. A multivariate method for dichotomous outcome variables.
- Hotelling’s T2. The 0/1 outcome is turned into the grouping variable, and the former predictors are turned into outcome variables. This will produce an overall test of significance but will not give individual coefficients for each variable, and it is unclear the extent to which each “predictor” is adjusted for the impact of the other “predictors”.
Using the Probit Model
The code below estimates a probit regression model using the glm (generalized linear model) function.
Since we stored our model output in the object “myprobit”, R will not print anything to the console. We
can use the summary function to get a summary of the
model and all the estimates.
myprobit <- glm(admit ~ gre + gpa + rank, family = binomial(link = "probit"), data = mydata) ## model summary summary(myprobit)
## ## Call: ## glm(formula = admit ~ gre + gpa + rank, family = binomial(link = "probit"), ## data = mydata) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -1.616 -0.871 -0.639 1.156 2.103 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -2.38684 0.67395 -3.54 0.00040 *** ## gre 0.00138 0.00065 2.12 0.03433 * ## gpa 0.47773 0.19720 2.42 0.01541 * ## rank2 -0.41540 0.19498 -2.13 0.03313 * ## rank3 -0.81214 0.20836 -3.90 9.7e-05 *** ## rank4 -0.93590 0.24527 -3.82 0.00014 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## ## Null deviance: 499.98 on 399 degrees of freedom ## Residual deviance: 458.41 on 394 degrees of freedom ## AIC: 470.4 ## ## Number of Fisher Scoring iterations: 4
- In the output above, the first thing we see is the call, this is R reminding us what the model we ran was, what options we specified, etc.
- Next we see the deviance residuals, which are a measure of model fit. This part of output shows the distribution of the deviance residuals for individual cases used in the model. Below we discuss how to use summaries of the deviance statistic to asses model fit.
- The next part of the output shows the coefficients, their standard errors,
the z-statistic (sometimes called a Wald z-statistic), and the associated
p-values. Both
gre,gpa, and the three terms forrankare statistically significant. The probit regression coefficients give the change in the z-score or probit index for a one unit change in the predictor.- For a one unit increase in
gre, the z-score increases by 0.001. - For each one unit increase in
gpa, the z-score increases by 0.478. - The indicator variables for
rankhave a slightly different interpretation. For example, having attended an undergraduate institution ofrankof 2, versus an institution with arankof 1 (the reference group), decreases the z-score by 0.415.
- For a one unit increase in
- Below the table of coefficients are fit indices, including the null and deviance residuals and the AIC. Later we show an example of how you can use these values to help assess model fit.
We can use the confint function to obtain confidence intervals for the coefficient estimates.
These will be profiled confidence intervals by default, created by profiling the likelihood function. As
such, they are not necessarily symmetric.
confint(myprobit)
## 2.5 % 97.5 % ## (Intercept) -3.7201051 -1.076328 ## gre 0.0001104 0.002655 ## gpa 0.0960655 0.862610 ## rank2 -0.7992114 -0.032995 ## rank3 -1.2230956 -0.405008 ## rank4 -1.4234218 -0.459539
We can test for an overall effect of rank using the wald.test
function of the aod library. The
order in which the coefficients are given in the table of coefficients is the
same as the order of the terms in the model. This is important because the
wald.test function refers to the coefficients by their order in
the model.
We use the wald.test function. b
supplies the coefficients, while Sigma supplies the variance covariance
matrix of the error terms, finally Terms tells R which terms in the model
are to be tested, in this case, terms 4, 5, and 6, are the three terms for the
levels of rank.
wald.test(b = coef(myprobit), Sigma = vcov(myprobit), Terms = 4:6)
## Wald test: ## ---------- ## ## Chi-squared test: ## X2 = 21.4, df = 3, P(> X2) = 8.9e-05
The chi-squared test statistic of 21.4 with three degrees of freedom is
associated with a p-value of less than 0.001 indicating that the overall effect of
rank is statistically significant.
We can also test additional hypotheses about the differences in the
coefficients for different levels of rank. Below we
test that the coefficient for rank=2 is equal to the coefficient for rank=3.
The first line of code below creates a vector l that defines the test we
want to perform. In this case, we want to test the difference (subtraction) of
the terms for rank=2 and rank=3 (i.e. the 4th and 5th terms in the
model). To contrast these two terms, we multiply one of them by 1, and the other
by -1. The other terms in the model are not involved in the test, so they are
multiplied by 0. The second line of code below uses L=l to tell R that we
wish to base the test on the vector l (rather than using the Terms option
as we did above).
l <- cbind(0, 0, 0, 1, -1, 0) wald.test(b = coef(myprobit), Sigma = vcov(myprobit), L = l)
## Wald test: ## ---------- ## ## Chi-squared test: ## X2 = 5.6, df = 1, P(> X2) = 0.018
The chi-squared test statistic of 5.5 with 1 degree of freedom is associated with
a p-value of 0.019, indicating that the difference between the coefficient for rank=2
and the coefficient for rank=3 is statistically significant.
You can also use predicted probabilities to help you understand the model. To do this, we first create a data frame containing the values we want for the independent variables.
newdata <- data.frame(gre = rep(seq(from = 200, to = 800, length.out = 100), 4 * 4), gpa = rep(c(2.5, 3, 3.5, 4), each = 100 * 4), rank = factor(rep(rep(1:4, each = 100), 4))) head(newdata)
## gre gpa rank ## 1 200.0 2.5 1 ## 2 206.1 2.5 1 ## 3 212.1 2.5 1 ## 4 218.2 2.5 1 ## 5 224.2 2.5 1 ## 6 230.3 2.5 1
Now we can predict the probabilities for our input data as well as their standard errors.
These are stored as new variable in the data frame with the original data, so we can
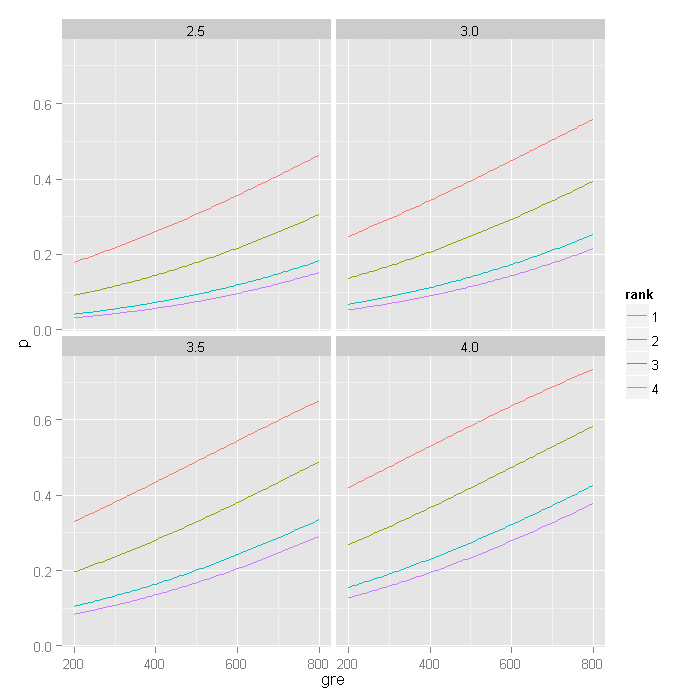
plot the predicted probabilities for different gre scores. We create four plots,
one for each level of gpa we used (2.5, 3, 3.5, 4) with the colour of the lines
indicating the rank the predicted probabilities were for.
newdata[, c("p", "se")] <- predict(myprobit, newdata, type = "response", se.fit = TRUE)[-3] ggplot(newdata, aes(x = gre, y = p, colour = rank)) + geom_line() + facet_wrap(~gpa)
We may also wish to see measures of how well our model fits. This can be
particularly useful when comparing competing models. The output produced by
summary(mylogit) included indices of fit (shown below the coefficients), including the null and
deviance residuals and the AIC. One measure of model fit is the significance of
the overall model. This test asks whether the model with predictors fits
significantly better than a model with just an intercept (i.e. a null model).
The test statistic is the difference between the residual deviance for the model
with predictors and the null model. The test statistic is distributed
chi-squared with degrees of freedom equal to the differences in degrees of freedom between
the current and the null model (i.e. the number of predictor variables in the
model). To find the difference in deviance for the two models (i.e. the test
statistic) we can compute the change in deviance, and test it using a chi square test—the
change in deviance distributed as chi square on the change in degrees
of freedom.
## change in deviance with(myprobit, null.deviance - deviance)
## [1] 41.56
## change in degrees of freedom with(myprobit, df.null - df.residual)
## [1] 5
## chi square test p-value with(myprobit, pchisq(null.deviance - deviance, df.null - df.residual, lower.tail = FALSE))
## [1] 7.219e-08
The chi-square of 41.56 with 5 degrees of freedom and an associated p-value of less than 0.001 tells us that our model as a whole fits significantly better than an empty model. This is sometimes called a likelihood ratio test (the deviance residual is -2*log likelihood). To see the model’s log likelihood, we type:
logLik(myprobit)
## 'log Lik.' -229.2 (df=6)
Things to consider
- Empty cells or small cells: You should check for empty or small cells by doing a crosstab between categorical predictors and the outcome variable. If a cell has very few cases (a small cell), the model may become unstable or it might not run at all.
- Separation or quasi-separation (also called perfect prediction), a condition in which the outcome does not vary at some levels of the independent variables. See our page FAQ: What is complete or quasi-complete separation in logistic/probit regression and how do we deal with them? for information on models with perfect prediction.
- Sample size: Both probit and logit models require more cases than OLS regression because they use maximum likelihood estimation techniques. It is sometimes possible to estimate models for binary outcomes in datasets with only a small number of cases using exact logistic regression. It is also important to keep in mind that when the outcome is rare, even if the overall dataset is large, it can be difficult to estimate a probit model.
- Pseudo-R-squared: Many different measures of psuedo-R-squared exist. They all attempt to provide information similar to that provided by R-squared in OLS regression; however, none of them can be interpreted exactly as R-squared in OLS regression is interpreted. For a discussion of various pseudo-R-squareds see Long and Freese (2006) or our FAQ page What are pseudo R-squareds?
- Diagnostics: The diagnostics for probit regression are different from those for OLS regression. The diagnostics for probit models are similar to those for logit models. For a discussion of model diagnostics for logistic regression, see Hosmer and Lemeshow (2000, Chapter 5).
References
- Hosmer, D. & Lemeshow, S. (2000). Applied Logistic Regression (Second Edition). New York: John Wiley & Sons, Inc.
- Long, J. Scott (1997). Regression Models for Categorical and Limited Dependent Variables. Thousand Oaks, CA: Sage Publications.
See Also
- R Online Manual: glm
- Applied Logistic Regression (Second Edition) by David Hosmer and Stanley Lemeshow
- Stat Books for Loan, Logistic Regression and Limited Dependent Variables
- Everitt, B. S. and Hothorn, T. A Handbook of Statistical Analyses Using R
