The R package(s) needed for this chapter is the survival package. We currently use R 2.0.1 patched version. You may want to make sure that packages on your local machine are up to date. You can perform updating in R using update.packages() function.
Table 1.1 on page 4, data set is hmohiv.csv.
hmohiv<-read.table("https://stats.idre.ucla.edu/stat/r/examples/asa/hmohiv.csv", sep=",", header = TRUE)
attach(hmohiv)
hmohiv
ID time age drug censor entdate enddate 1 1 5 46 0 1 5/15/1990 10/14/1990 2 2 6 35 1 0 9/19/1989 3/20/1990 3 3 8 30 1 1 4/21/1991 12/20/1991 4 4 3 30 1 1 1/3/1991 4/4/1991 5 5 22 36 0 1 9/18/1989 7/19/1991 6 6 1 32 1 0 3/18/1991 4/17/1991 7 7 7 36 1 1 11/11/1989 6/11/1990 8 8 9 31 1 1 11/25/1989 8/25/1990 9 9 3 48 0 1 2/11/1991 5/13/1991 10 10 12 47 0 1 8/11/1989 8/11/1990 ...........(part of output omitted here)........... 91 91 4 35 0 1 4/10/1990 8/9/1990 92 92 57 36 0 1 12/11/1990 9/9/1995 93 93 1 41 1 1 12/15/1990 1/14/1991 94 94 12 36 1 0 1/13/1989 1/13/1990 95 95 7 35 1 1 8/22/1991 3/21/1992 96 96 1 34 1 1 8/2/1991 9/1/1991 97 97 5 28 0 1 5/22/1991 10/21/1991 98 98 60 29 0 0 4/2/1990 4/1/1995 99 99 2 35 1 0 5/1/1991 6/30/1991 100 100 1 34 1 1 5/11/1989 6/10/1989
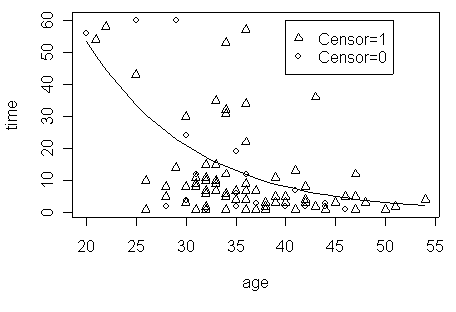
Figure 1.1 on page 6 using the hmohiv data set. To control the type of symbol, a variable called psymbol is created. It takes value 1 and 2, so the symbol type will be 1 and 2.
psymbol<-censor+1
table(psymbol)
psymbol
1 2
20 80
plot(age, time, pch=(psymbol))
legend(40, 60, c("Censor=1", "Censor=0"), pch=(psymbol))
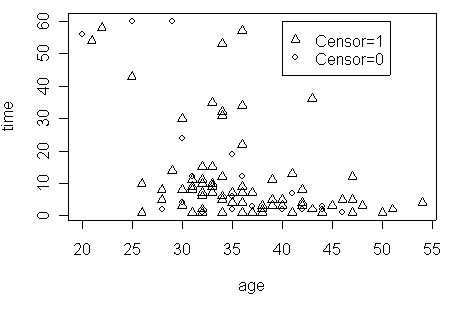
Figure 1.2 on page 7 using the hmohiv data set.
age1<-1000/age
plot(age1, time, pch=(psymbol))
legend(40, 30, c("Censor=1", "Censor=0"), pch=(psymbol))
Table 1.2 on page 14 using the data set hmohiv. Package “survival” is needed for this analysis and for most of the analyses in the book.
library(survival)
test
Call:
survreg(formula = Surv(time, censor) ~ age, dist = "exponential")
Value Std. Error z p
(Intercept) 5.859 0.5853 10.01 1.37e-23
age -0.094 0.0158 -5.96 2.59e-09
Scale fixed at 1
Exponential distribution
Loglik(model)= -275 Loglik(intercept only)= -292.3
Chisq= 34.5 on 1 degrees of freedom, p= 4.3e-09
Number of Newton-Raphson Iterations: 4
n= 100
Figure 1.3 on page 16 using data set hmohiv and the model created for Table 1.2 in previous example.
pred