The R package(s) needed for this chapter is the survival package. We currently use R 2.0.1 patched version. You may want to make sure that packages on your local machine are up to date. You can perform updating in R using update.packages() function.
Table 5.1 on page 166 using data set uis on different covariates.
- Variable hercoc:
rm(list=ls())
library(survival)
uis<-read.table("https://stats.idre.ucla.edu/stat/r/examples/asa/uis.csv", sep=",", header = TRUE)
attach(uis)
hercoc.ph <- coxph( Surv(time, censor) ~ factor(hercoc), method="breslow") summary(hercoc.ph)
n=610 (18 observations deleted due to missing)
coef exp(coef) se(coef) z p
factor(hercoc)2 0.0773 1.080 0.144 0.535 0.59
factor(hercoc)3 -0.2544 0.775 0.135 -1.883 0.06
factor(hercoc)4 -0.1617 0.851 0.130 -1.243 0.21
exp(coef) exp(-coef) lower .95 upper .95 factor(hercoc)2 1.080 0.926 0.814 1.43 factor(hercoc)3 0.775 1.290 0.595 1.01 factor(hercoc)4 0.851 1.176 0.659 1.10
Rsquare= 0.013 (max possible= 1 ) Likelihood ratio test= 7.76 on 3 df, p=0.0513 Wald test = 7.88 on 3 df, p=0.0486 Score (logrank) test = 7.92 on 3 df, p=0.0476
survfit( Surv(time, censor) ~ hercoc, uis)
18 observations deleted due to missing
n events median 0.95LCL 0.95UCL
hercoc=1 111 92 150 107 198
hercoc=2 114 100 147 110 184
hercoc=3 178 136 185 148 231
hercoc=4 207 165 181 155 220
- Variable ivhx:
detach()
ivhx.ph<-coxph( Surv(time, censor) ~ factor(ivhx), data=uis, method="breslow") summary(ivhx.ph)
n=610 (18 observations deleted due to missing)
coef exp(coef) se(coef) z p
factor(ivhx)2 0.195 1.22 0.129 1.52 0.13000
factor(ivhx)3 0.385 1.47 0.101 3.80 0.00014
exp(coef) exp(-coef) lower .95 upper .95 factor(ivhx)2 1.22 0.822 0.945 1.56 factor(ivhx)3 1.47 0.680 1.205 1.79
Rsquare= 0.024 (max possible= 1 ) Likelihood ratio test= 14.6 on 2 df, p=0.000663 Wald test = 14.5 on 2 df, p=0.000705 Score (logrank) test = 14.7 on 2 df, p=0.000654
survfit( Surv(time, censor) ~ ivhx, uis)
18 observations deleted due to missing
n events median 0.95LCL 0.95UCL
ivhx=1 233 173 194 175 231
ivhx=2 115 93 170 130 231
ivhx=3 262 227 148 115 168
- Variable race:
race.ph<-coxph( Surv(time, censor) ~ factor(race), uis, method="breslow") summary(race.ph)
n=622 (6 observations deleted due to missing)
coef exp(coef) se(coef) z p
factor(race)1 -0.284 0.753 0.106 -2.68 0.0073
exp(coef) exp(-coef) lower .95 upper .95 factor(race)1 0.753 1.33 0.611 0.926
Rsquare= 0.012 (max possible= 1 ) Likelihood ratio test= 7.57 on 1 df, p=0.00593 Wald test = 7.21 on 1 df, p=0.00726 Score (logrank) test = 7.26 on 1 df, p=0.00706
stci.hl(50, uis, subset=race==0)
quantile time cie.lower cie.upper
[1,] 50 152 124 174
stci.hl(50, uis, subset=race==1)
quantile time cie.lower cie.upper
[1,] 50 193 162 232
- Variable treat:
treat.ph<-coxph( Surv(time, censor) ~ factor(treat), uis, method="breslow") summary(treat.ph)
n= 628
coef exp(coef) se(coef) z p
factor(treat)1 -0.231 0.794 0.089 -2.60 0.0094
exp(coef) exp(-coef) lower .95 upper .95 factor(treat)1 0.794 1.26 0.667 0.945
Rsquare= 0.011 (max possible= 1 ) Likelihood ratio test= 6.75 on 1 df, p=0.00937 Wald test = 6.74 on 1 df, p=0.00941 Score (logrank) test = 6.77 on 1 df, p=0.00926
survfit( Surv(time, censor) ~ treat, uis)
n events median 0.95LCL 0.95UCL
treat=0 320 265 132 115 156
treat=1 308 243 192 175 226
Variable site:
site.ph<-coxph( Surv(time, censor) ~ factor(site), uis, method="breslow") summary(site.ph)
n= 628
coef exp(coef) se(coef) z p
factor(site)1 -0.151 0.86 0.0986 -1.53 0.13
exp(coef) exp(-coef) lower .95 upper .95 factor(site)1 0.86 1.16 0.709 1.04
Rsquare= 0.004 (max possible= 1 ) Likelihood ratio test= 2.4 on 1 df, p=0.121 Wald test = 2.35 on 1 df, p=0.125 Score (logrank) test = 2.36 on 1 df, p=0.125
survfit( Surv(time, censor) ~ site, uis)
n events median 0.95LCL 0.95UCL
site=0 444 364 156 132 175
site=1 184 144 199 161 232
Variable age:
uis$agecat<-cut(uis$age, c(19, 27, 32,37,56)) agecat.ph<-coxph( Surv(time, censor) ~ agecat, uis, method="breslow") summary(agecat.ph)
n=623 (5 observations deleted due to missing)
coef exp(coef) se(coef) z p
factor(agecat)(27,32] 0.0808 1.084 0.123 0.654 0.51
factor(agecat)(32,37] -0.0658 0.936 0.121 -0.542 0.59
factor(agecat)(37,56] -0.1684 0.845 0.135 -1.247 0.21
exp(coef) exp(-coef) lower .95 upper .95 factor(agecat)(27,32] 1.084 0.922 0.851 1.38 factor(agecat)(32,37] 0.936 1.068 0.738 1.19 factor(agecat)(37,56] 0.845 1.183 0.649 1.10
Rsquare= 0.006 (max possible= 1 ) Likelihood ratio test= 3.81 on 3 df, p=0.282 Wald test = 3.79 on 3 df, p=0.285 Score (logrank) test = 3.8 on 3 df, p=0.284
agecat.surv<-survfit( Surv(time, censor) ~ agecat, uis)
agecat.surv
5 observations deleted due to missing
n events median 0.95LCL 0.95UCL
agecat=(19,27] 158 128 162 122 203
agecat=(27,32] 158 135 148 123 182
agecat=(32,37] 184 145 163 124 216
agecat=(37,56] 123 96 189 168 243
Variable becktoa:
uis$beckt2<-cut(uis$becktota, c(-.5, 9.99,14.9,24.9,55)) beckcat.ph<-coxph( Surv(time, censor) ~ beckt2, uis, method="breslow") summary(beckcat.ph)
n=595 (33 observations deleted due to missing)
coef exp(coef) se(coef) z p
beckt2(9.99,14.9] 0.0752 1.08 0.150 0.502 0.620
beckt2(14.9,24.9] 0.1821 1.20 0.123 1.485 0.140
beckt2(24.9,55] 0.2631 1.30 0.137 1.927 0.054
exp(coef) exp(-coef) lower .95 upper .95 beckt2(9.99,14.9] 1.08 0.928 0.804 1.45 beckt2(14.9,24.9] 1.20 0.834 0.943 1.53 beckt2(24.9,55] 1.30 0.769 0.996 1.70
Rsquare= 0.007 (max possible= 1 ) Likelihood ratio test= 4.41 on 3 df, p=0.221 Wald test = 4.37 on 3 df, p=0.225 Score (logrank) test = 4.38 on 3 df, p=0.223
beckcat.surv<-survfit( Surv(time, censor) ~ beckt2, uis, na.action=na.exclude)
beckcat.surv
33 observations deleted due to missing
n events median 0.95LCL 0.95UCL
beckt2=(-0.5,9.99] 135 104 211 170 248
beckt2=(9.99,14.9] 102 78 170 133 232
beckt2=(14.9,24.9] 226 185 168 139 200
beckt2=(24.9,55] 132 111 146 115 193
Variable ndurgtx:
uis$drugcat<-cut(uis$ndrugtx, c(-.5, 1, 3, 6, 70)) drugcat.ph<-coxph( Surv(time, censor) ~ drugcat, uis, method="breslow") summary(drugcat.ph)
n=611 (17 observations deleted due to missing)
coef exp(coef) se(coef) z p
drugcat(1,3] -0.0509 0.95 0.123 -0.412 0.680
drugcat(3,6] 0.2661 1.30 0.125 2.134 0.033
drugcat(6,70] 0.3649 1.44 0.127 2.880 0.004
exp(coef) exp(-coef) lower .95 upper .95 drugcat(1,3] 0.95 1.052 0.746 1.21 drugcat(3,6] 1.30 0.766 1.022 1.67 drugcat(6,70] 1.44 0.694 1.124 1.85
Rsquare= 0.023 (max possible= 1 ) Likelihood ratio test= 14.5 on 3 df, p=0.0023 Wald test = 14.7 on 3 df, p=0.00206 Score (logrank) test = 14.9 on 3 df, p=0.00192
drugcat.surv<-survfit( Surv(time, censor) ~ drugcat, uis, na.action=na.exclude) drugcat.surv Call: survfit(formula = Surv(time, censor) ~ drugcat, data = uis, na.action = na.exclude)
17 observations deleted due to missing
n events median 0.95LCL 0.95UCL
drugcat=(-0.5,1] 183 141 170 143 227
drugcat=(1,3] 165 123 177 162 211
drugcat=(3,6] 138 119 132 106 187
drugcat=(6,70] 125 113 123 110 186
Table 5.2 on page 167 using uis data set.
uis$age5 <- uis$age/5
uis$beck10 <- uis$becktota/10
uis$drug5 <- uis$ndrugtx/5
age5.ph<-coxph( Surv(time, censor) ~ age5, uis, method="breslow")
summary(age5.ph)
n=623 (5 observations deleted due to missing values)
coef exp(coef) se(coef) z p
age5 -0.0643 0.938 0.0359 -1.79 0.074
exp(coef) exp(-coef) lower .95 upper .95
age5 0.938 1.07 0.874 1.01
Rsquare= 0.005 (max possible= 1 )
Likelihood ratio test= 3.24 on 1 df, p=0.0719
Wald test = 3.2 on 1 df, p=0.0735
Score (logrank) test = 3.2 on 1 df, p=0.0735
beck10.ph<-coxph( Surv(time, censor) ~ beck10, uis, method="breslow")
summary(beck10.ph)
n=595 (33 observations deleted due to missing values)
coef exp(coef) se(coef) z p
beck10 0.11 1.12 0.0472 2.32 0.02
exp(coef) exp(-coef) lower .95 upper .95
beck10 1.12 0.896 1.02 1.22
Rsquare= 0.009 (max possible= 1 )
Likelihood ratio test= 5.32 on 1 df, p=0.0211
Wald test = 5.4 on 1 df, p=0.0201
Score (logrank) test = 5.41 on 1 df, p=0.0201
drug5.ph<-coxph( Surv(time, censor) ~ drug5, uis, method="breslow")
summary(drug5.ph)
n=611 (17 observations deleted due to missing)
coef exp(coef) se(coef) z p
drug5 0.147 1.16 0.0375 3.92 9e-05
exp(coef) exp(-coef) lower .95 upper .95 drug5 1.16 0.863 1.08 1.25
Rsquare= 0.022 (max possible= 1 ) Likelihood ratio test= 13.4 on 1 df, p=0.000259 Wald test = 15.4 on 1 df, p=8.95e-05 Score (logrank) test = 15.5 on 1 df, p=8.44e-05
Table 5.3 on page 168 using data set uis.
full.ph <- coxph( Surv(time, censor) ~ age + becktota + ndrugtx + factor(hercoc)
+ factor(ivhx) + race + treat + site, uis, method="breslow")
summary(full.ph)
n=575 (53 observations deleted due to missing)
coef exp(coef) se(coef) z p
age -0.02887 0.972 0.00817 -3.533 0.00041
becktota 0.00834 1.008 0.00498 1.677 0.09400
ndrugtx 0.02837 1.029 0.00831 3.416 0.00064
factor(hercoc)2 0.06532 1.067 0.15001 0.435 0.66000
factor(hercoc)3 -0.09362 0.911 0.16547 -0.566 0.57000
factor(hercoc)4 0.02798 1.028 0.16028 0.175 0.86000
factor(ivhx)2 0.17439 1.191 0.13864 1.258 0.21000
factor(ivhx)3 0.28071 1.324 0.14693 1.911 0.05600
race -0.20289 0.816 0.11669 -1.739 0.08200
treat -0.23995 0.787 0.09437 -2.543 0.01100
site -0.10249 0.903 0.10927 -0.938 0.35000
exp(coef) exp(-coef) lower .95 upper .95 age 0.972 1.029 0.956 0.987 becktota 1.008 0.992 0.999 1.018 ndrugtx 1.029 0.972 1.012 1.046 factor(hercoc)2 1.067 0.937 0.796 1.432 factor(hercoc)3 0.911 1.098 0.658 1.259 factor(hercoc)4 1.028 0.972 0.751 1.408 factor(ivhx)2 1.191 0.840 0.907 1.562 factor(ivhx)3 1.324 0.755 0.993 1.766 race 0.816 1.225 0.649 1.026 treat 0.787 1.271 0.654 0.946 site 0.903 1.108 0.729 1.118
Rsquare= 0.08 (max possible= 1 ) Likelihood ratio test= 47.9 on 11 df, p=1.48e-06 Wald test = 48.8 on 11 df, p=1.04e-06 Score (logrank) test = 49.4 on 11 df, p=8.17e-07
Table 5.4 on page 168 using data set uis with smaller model comparing with the previous example.
reduced1.ph <- coxph( Surv(time, censor) ~ age + becktota + ndrugtx +
factor(ivhx) + race + treat + site, uis, method="breslow")
summary(reduced1.ph)
n=575 (53 observations deleted due to missing)
coef exp(coef) se(coef) z p
age -0.02822 0.972 0.00817 -3.454 0.00055
becktota 0.00794 1.008 0.00497 1.598 0.11000
ndrugtx 0.02776 1.028 0.00829 3.350 0.00081
factor(ivhx)2 0.19599 1.217 0.13721 1.428 0.15000
factor(ivhx)3 0.33280 1.395 0.11991 2.775 0.00550
race -0.20925 0.811 0.11589 -1.805 0.07100
treat -0.23177 0.793 0.09371 -2.473 0.01300
site -0.09946 0.905 0.10854 -0.916 0.36000
exp(coef) exp(-coef) lower .95 upper .95 age 0.972 1.029 0.957 0.988 becktota 1.008 0.992 0.998 1.018 ndrugtx 1.028 0.973 1.012 1.045 factor(ivhx)2 1.217 0.822 0.930 1.592 factor(ivhx)3 1.395 0.717 1.103 1.764 race 0.811 1.233 0.646 1.018 treat 0.793 1.261 0.660 0.953 site 0.905 1.105 0.732 1.120
Rsquare= 0.078 (max possible= 1 ) Likelihood ratio test= 46.5 on 8 df, p=1.90e-07 Wald test = 47.5 on 8 df, p=1.24e-07 Score (logrank) test = 48 on 8 df, p=9.92e-08
Table 5.5 using data set uis, a further reduced model based on the previous example.
uis$ivhx3<-(uis$ivhx==3)
reduced2.ph <- coxph( Surv(time, censor) ~ age + becktota + ndrugtx +
ivhx3 + race + treat + site, uis, method="breslow")
summary(reduced2.ph)
n=575 (53 observations deleted due to missing)
coef exp(coef) se(coef) z p
age -0.0262 0.974 0.00805 -3.249 0.0012
becktota 0.0084 1.008 0.00495 1.696 0.0900
ndrugtx 0.0291 1.030 0.00821 3.540 0.0004
ivhx3TRUE 0.2561 1.292 0.10630 2.409 0.0160
race -0.2245 0.799 0.11527 -1.947 0.0510
treat -0.2324 0.793 0.09373 -2.480 0.0130
site -0.0867 0.917 0.10786 -0.804 0.4200
exp(coef) exp(-coef) lower .95 upper .95 age 0.974 1.026 0.959 0.990 becktota 1.008 0.992 0.999 1.018 ndrugtx 1.030 0.971 1.013 1.046 ivhx3TRUE 1.292 0.774 1.049 1.591 race 0.799 1.252 0.637 1.001 treat 0.793 1.262 0.660 0.952 site 0.917 1.091 0.742 1.133
Rsquare= 0.074 (max possible= 1 ) Likelihood ratio test= 44.5 on 7 df, p=1.7e-07 Wald test = 45.5 on 7 df, p=1.07e-07 Score (logrank) test = 46 on 7 df, p=8.67e-08
Table 5.6 using data set uis based on the model created in the previous example and the categorical variables created for Table 5.1.
uis$agecat<-cut(uis$age, c(19, 27, 32,37,56))
agecat.ph <- coxph(Surv(time, censor) ~ agecat + becktota + ndrugtx +
ivhx3 + site + race + treat, uis, method="breslow")
summary(agecat.ph)
n=575 (53 observations deleted due to missing)
coef exp(coef) se(coef) z p
agecat(27,32] 0.03582 1.036 0.13126 0.273 0.78000
agecat(32,37] -0.20940 0.811 0.13003 -1.610 0.11000
agecat(37,56] -0.39059 0.677 0.14996 -2.605 0.00920
becktota 0.00881 1.009 0.00493 1.787 0.07400
ndrugtx 0.02920 1.030 0.00832 3.510 0.00045
ivhx3TRUE 0.24242 1.274 0.10634 2.280 0.02300
site -0.07911 0.924 0.10849 -0.729 0.47000
race -0.24549 0.782 0.11618 -2.113 0.03500
treat -0.22935 0.795 0.09403 -2.439 0.01500
(output omitted.)
uis$beckcat<-cut(uis$becktota, c(-.5, 9.99,14.9,24.9,55))
beckcat.ph <- coxph(Surv(time, censor) ~ age + beckcat + ndrugtx +
ivhx3 + site + race + treat, uis, method="breslow")
summary(beckcat.ph)
n=575 (53 observations deleted due to missing)
coef exp(coef) se(coef) z p
age -0.0265 0.974 0.00804 -3.297 0.00098
beckcat(9.99,14.9] 0.0695 1.072 0.15212 0.457 0.65000
beckcat(14.9,24.9] 0.0909 1.095 0.12493 0.728 0.47000
beckcat(24.9,55] 0.2169 1.242 0.13995 1.550 0.12000
ndrugtx 0.0289 1.029 0.00825 3.509 0.00045
ivhx3TRUE 0.2616 1.299 0.10631 2.461 0.01400
site -0.0920 0.912 0.10782 -0.853 0.39000
race -0.2254 0.798 0.11549 -1.952 0.05100
treat -0.2304 0.794 0.09390 -2.453 0.01400
(output omitted)
uis$drugcat<-cut(ndrugtx, c(-.5, 1, 3, 6, 70))
drugcat.ph <- coxph(Surv(time, censor) ~ age + becktota + drugcat +
ivhx3 + site + race + treat, uis, method="breslow")
summary(drugcat.ph)
n=575 (53 observations deleted due to missing)
coef exp(coef) se(coef) z p
age -0.02772 0.973 0.00815 -3.402 0.00067
becktota 0.00805 1.008 0.00495 1.626 0.10000
drugcat(1,3] -0.06951 0.933 0.12911 -0.538 0.59000
drugcat(3,6] 0.25949 1.296 0.13368 1.941 0.05200
drugcat(6,70] 0.39935 1.491 0.14030 2.846 0.00440
ivhx3TRUE 0.24908 1.283 0.10664 2.336 0.02000
site -0.08384 0.920 0.10791 -0.777 0.44000
race -0.22966 0.795 0.11542 -1.990 0.04700
treat -0.21991 0.803 0.09357 -2.350 0.01900
(output omitted)
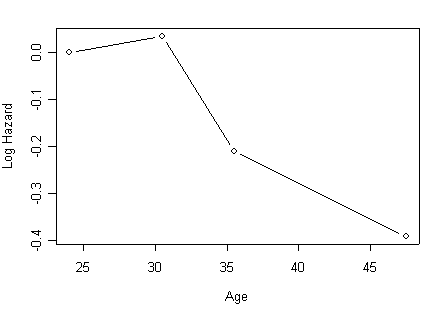
Figure 5.1 (a), (b) and (c) based on the models from Table 5.6.
Panel (a) Estimated coefficients for grouped age using the object agecat.ph created in the previous example.
names(agecat.ph)
[1] "coefficients" "var" "loglik"
[4] "score" "iter" "linear.predictors"
[7] "residuals" "means" "method"
[10] "n" "terms" "assign"
[13] "wald.test" "na.action" "y"
[16] "formula" "call"
agecat.ph$coefficients
agecat(27,32] agecat(32,37] agecat(37,56] becktota ndrugtx
0.03582336 -0.20940371 -0.39058778 0.00881022 0.02919603
ivhx3TRUE site race treat
0.24241703 -0.07910672 -0.24548561 -0.22934666
age.coeff<-data.frame(agecat.ph$coefficients)[1:3,]
age.plot<-cbind(midpt=c(24, 30.5, 35.5, 47.5), rbind(0, data.frame(age.coeff)))
age.plot
midpt age.coeff
1 24.0 0.00000000
11 30.5 0.03582336
2 35.5 -0.20940371
3 47.5 -0.39058778
plot(age.plot[,1], age.plot[,2], ylab="Log Hazard", xlab="Age", type="b")
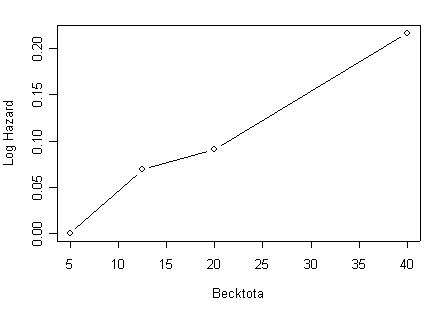
Panel (b) Estimated coefficients for grouped beck score using the object beckcat.ph created in the previous example.
beckcat.ph$coefficients
age beckcat(9.99,14.9] beckcat(14.9,24.9] beckcat(24.9,55]
-0.02650867 0.06948882 0.09094400 0.21687651
ndrugtx ivhx3TRUE site race
0.02894029 0.26163192 -0.09197744 -0.22540640
treat
-0.23038130
beck.coeff<-data.frame(beckcat.ph$coefficients)[2:4,]
beck.plot<-cbind(midpt=c(5, 12.5, 20, 40), rbind(0, data.frame(beck.coeff)))
plot(beck.plot[,1], beck.plot[,2], ylab="Log Hazard", xlab="Becktota", type="b")
Panel (c) Estimated coefficients for grouped drug treatments using the object drugcat.ph created in the previous example.
drugcat.ph$coefficients
age becktota drugcat(1,3] drugcat(3,6] drugcat(6,70]
-0.027723404 0.008050683 -0.069508432 0.259486831 0.399345037
ivhx3TRUE site race treat
0.249076482 -0.083840409 -0.229664884 -0.219909556
drug.coeff<-data.frame(drugcat.ph$coefficients)[3:5,]
drug.plot<-cbind(midpt=c(.5, 2.5, 5.0, 23.5), rbind(0, data.frame(drug.coeff)))
drug.plot
midpt drug.coeff
1 0.5 0.00000000
11 2.5 -0.06950843
2 5.0 0.25948683
3 23.5 0.39934504
plot(drug.plot[,1], drug.plot[,2], ylab="Log Hazard", xlab="NDRUGTX", type="b")
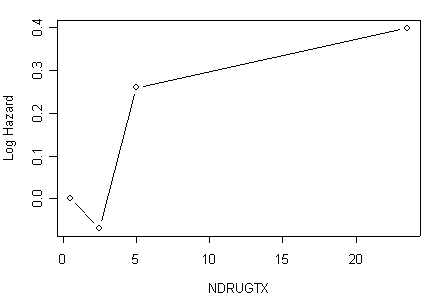
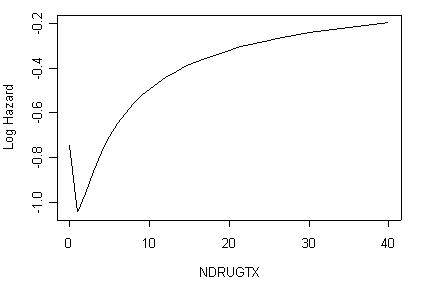
Figure 5.1 (d) on page 171 and Table 5.7 on page 172 using data set uis and fractional polynomials for ndrugtx. Currently (3/18/05), package mfp does multiple fractional polynomials in regression models, including cox models. You can download the package by “install.packages(“mfp”)” assuming that your machine is connected to the internet. Unlike Stata’s fracpoly command, function mfp in R does not automatically transform variables into desirable forms. For example, if a variable takes value zero sometimes, then negative power is not being used, thus restricting some possible better transformation. In this example, variable ndrugtx takes value zero sometimes, we will apply mfp on a transformed variable.
rm(list=ls())
library(survival)
library(mfp)
uis<-read.table("https://stats.idre.ucla.edu/stat/r/examples/asa/uis.csv", sep=",", header = TRUE)
uis$ivhx3<-(uis$ivhx==3)
uis$nx<-10/(uis$ndrugtx+1)
attach(uis)
uis$ok<-complete.cases(uis$age, uis$becktota, uis$nx, uis$ivhx3, uis$site, uis$race, uis$treat)
to.uis<-uis[ok,]
# Not in the mdoel
a.ph<- coxph(Surv(time, censor) ~ age + becktota + ivhx3 + site + race + treat,
to.uis, method="breslow")
2*a.ph$loglik [1] -5327.970 -5294.497 # Linear term
alin<- coxph(Surv(time, censor) ~ ndrugtx + age+ becktota + ivhx3 + site + race + treat,
to.uis, method="breslow")
2*alin$loglik
[1] -5327.970 -5283.459
# J = 1 (2 df)
a2<- mfp(Surv(time, censor) ~fp(nx, df=2) + age+ becktota + ivhx3 + site + race + treat,
family = cox, data = to.uis)
a2$dev
[1] 5281.24
a2$powers
power1 power2
site 1.0 NA
nx -0.5 NA
becktota 1.0 NA
race 1.0 NA
treat 1.0 NA
ivhx3TRUE 1.0 NA
age 1.0 NA
# J = 2 (4 df)
a4<- mfp(Surv(time, censor) ~fp(nx, df=4) + age+ becktota + ivhx3 + site + race + treat,
family = cox, data = to.uis)
a4$powers
power1 power2
site 1 NA
nx 1 1
becktota 1 NA
race 1 NA
treat 1 NA
ivhx3TRUE 1 NA
age 1 NA
a4$dev [1] 5274.678
Now we are ready to create the plot (d) of Figure 5.1 based on object a4.
names(a4) [1] "coefficients" "var" "loglik" [4] "score" "iter" "linear.predictors" [7] "residuals" "means" "method" [10] "x" "powers" "pvalues" [13] "scale" "df.initial" "df.final" [16] "dev" "dev.lin" "dev.null" [19] "fptable" "n" "family" [22] "wald.test" "X" "y" [25] "terms" "call" head(a4$x) site.1 nx.1 nx.2 becktota.1 race.1 treat.1 ivhx3TRUE.1 age.1 1 0 5.000000 8.0471896 9.00 0 1 1 39 2 0 1.111111 0.1170672 34.00 0 1 0 33 3 0 2.500000 2.2907268 10.00 0 1 1 33 4 0 5.000000 8.0471896 20.00 0 0 1 32 5 0 1.666667 0.8513760 5.00 1 1 0 24 6 0 5.000000 8.0471896 32.55 0 1 1 30
y<-a4$x[,2]*(-0.52360) + a4$x[,3]*0.19505 f51.d<-cbind(x=to.uis$ndrugtx, y) f51.d<-f51.d[order(f51.d$x),] par(cex=.8) plot(f51.d$x, f51.d$y, type="l", xlab="NDRUGTX", ylab="Log Hazard")
Table 5.8 on page 174 based on the fractional polynomial model in previous example.
a4
Call:
mfp(formula = Surv(time, censor) ~ fp(nx, df = 4) + age + becktota +
ivhx3 + site + race + treat, data = to.uis, family = cox)
coef exp(coef) se(coef) z p site.1 -0.10583 0.900 0.10916 -0.97 3.3e-01 nx.1 -0.52360 0.592 0.12441 -4.21 2.6e-05 nx.2 0.19505 1.215 0.04825 4.04 5.3e-05 becktota.1 0.00918 1.009 0.00499 1.84 6.6e-02 race.1 -0.24242 0.785 0.11547 -2.10 3.6e-02 treat.1 -0.21138 0.809 0.09369 -2.26 2.4e-02 ivhx3TRUE.1 0.25911 1.296 0.10802 2.40 1.6e-02 age.1 -0.02820 0.972 0.00813 -3.47 5.2e-04
Likelihood ratio test=51.6 on 8 df, p=1.99e-08 n= 575
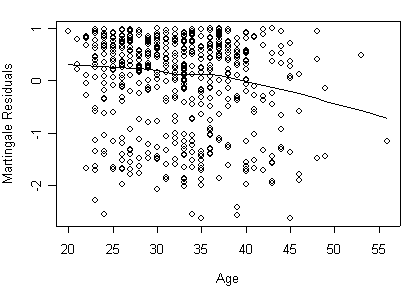
Figure 5.2 on page 175 with Martingale residuals and Lowess smoothed residuals. Library Design includes functions for computing different types of residuals for survival models. You may want have to download the package first by install.packages(“Design”).
Panel (a) Martingale residuals and Lowess smoothed residuals.
detach(uis)
attach(to.uis)
age.ph <- coxph( Surv(time, censor) ~ becktota + ndrugtx + ivhx3
+ race + treat + site, to.uis, method="breslow")
to.uis$resid<- residuals(age.ph, type="martingale", data=to.uis)
plot(to.uis$age, to.uis$resid, xlab="Age",ylab="Martingale Residuals")
lines(lowess(to.uis$age, to.uis$resid))
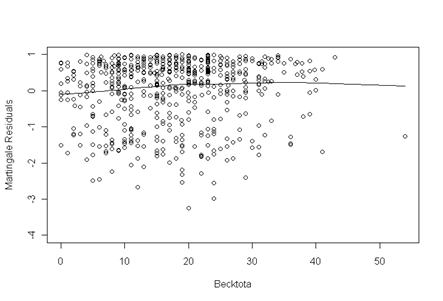
Panel (b) Log (Smoothed Censor/Smoothed Expected) + Beta*age.
Fig. 5.3a, p. 176. The fitted line appears to be approximately linear thus there is no transformation of becktota needed.
becktota.ph <- coxph( Surv(time, censor) ~ age + ndrugtx +
ivhx3 + race + treat + site, uis, method="breslow", na.action=na.exclude)
uis$resid <- residuals(becktota.ph, type="martingale")
plot(uis$becktota, uis$resid, xlab="Becktota",ylab="Martingale Residuals", ylim=c(-4, 1.0))
lines(lowess(uis$becktota[!is.na(uis$becktota) & !is.na(uis$resid)],
uis$resid[!is.na(uis$becktota) & !is.na(uis$resid)]))
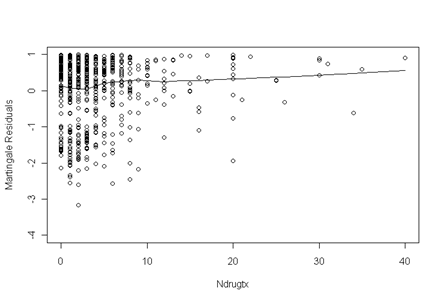
Fig. 5.4a, p. 177. The fitted line has a noticeable squiggle in the beginning which makes us very nervous and it is an indicator that we can’t just include ndrugtx in the model; we should transform it before including it in the final model.
ndrugtx.ph
Creating ndrugfp1 and ndrugfp2, p. 174.
uis$ndrugfp1 <- 1/((uis$ndrugtx+1)/10) uis$ndrugfp2 <- (1/((uis$ndrugtx+1)/10))*log((uis$ndrugtx+1)/10)
Table 5.8, p. 174.
reduced3.ph <- coxph( Surv(time, censor)~ age + becktota + ndrugfp1 + ndrugfp2 +
ivhx3 + race + treat + site, uis, method="breslow", na.action=na.exclude)
summary(reduced3.ph)
<output omitted>
n=575 (53 observations deleted due to missing values)
coef exp(coef) se(coef) z p
age -0.02815 0.972 0.00813 -3.461 0.000540
becktota 0.00916 1.009 0.00499 1.837 0.066000
ndrugfp1 -0.52284 0.593 0.12441 -4.202 0.000026
ndrugfp2 -0.19478 0.823 0.04825 -4.037 0.000054
ivhx3 0.25858 1.295 0.10802 2.394 0.017000
race -0.24220 0.785 0.11547 -2.098 0.036000
treat -0.21084 0.810 0.09369 -2.250 0.024000
site -0.10532 0.900 0.10915 -0.965 0.330000
Rsquare= 0.086 (max possible= 1 )
Likelihood ratio test= 51.4 on 8 df, p=2.17e-008
Wald test = 51.4 on 8 df, p=2.17e-008
Score (logrank) test = 52 on 8 df, p=1.72e-008
Creating the interaction variables, p. 178.
uis$agesite <- uis$age*uis$site uis$racesite <- uis$race*uis$site uis$agefp1 <- uis$age*uis$ndrugfp1 uis$agefp2 <- uis$age*uis$ndrugfp2
Table 5.10, p. 179.
interaction.ph <- coxph(formula=Surv(time, censor) ~ age + becktota + + ndrugfp1 + ndrugfp2 +
ivhx3 + race + treat + site + agesite + racesite + agefp1 + agefp2,
data=uis, method="breslow", na.action=na.exclude)
summary(interaction.ph)
<output omitted>
n=575 (53 observations deleted due to missing values)
coef exp(coef) se(coef) z p
age -0.05431 0.947 0.02803 -1.9372 0.05300
becktota 0.01005 1.010 0.00499 2.0129 0.04400
ndrugfp1 -0.67435 0.509 0.64447 -1.0464 0.30000
ndrugfp2 -0.17217 0.842 0.25234 -0.6823 0.50000
ivhx3 0.22935 1.258 0.10793 2.1250 0.03400
race -0.48774 0.614 0.13461 -3.6233 0.00029
treat -0.24205 0.785 0.09455 -2.5600 0.01000
site -1.11940 0.326 0.54607 -2.0499 0.04000
agesite 0.02644 1.027 0.01658 1.5949 0.11000
racesite 0.86274 2.370 0.24810 3.4773 0.00051
agefp1 0.00163 1.002 0.01945 0.0838 0.93000
agefp2 -0.00198 0.998 0.00766 -0.2589 0.80000
Rsquare= 0.119 (max possible= 1 )
Likelihood ratio test= 73.1 on 12 df, p=8.3e-011
Wald test = 73 on 12 df, p=8.61e-011
Score (logrank) test = 73.9 on 12 df, p=5.99e-011
Table 5.11, p. 179.
reducedinter.ph <- coxph(formula=Surv(time, censor) ~ age + becktota + + ndrugfp1 + ndrugfp2 +
ivhx3 + race + treat + site + agesite + racesite, uis, method="breslow", na.action=na.exclude)
summary(reducedinter.ph)
<output omitted>
n=575 (53 observations deleted due to missing values)
coef exp(coef) se(coef) z p
age -0.04138 0.959 0.00991 -4.17 3.0e-005
becktota 0.00874 1.009 0.00497 1.76 7.8e-002
ndrugfp1 -0.57473 0.563 0.12519 -4.59 4.4e-006
ndrugfp2 -0.21470 0.807 0.04859 -4.42 9.9e-006
ivhx3 0.22776 1.256 0.10857 2.10 3.6e-002
race -0.46661 0.627 0.13475 -3.46 5.3e-004
treat -0.24667 0.781 0.09434 -2.61 8.9e-003
site -1.31517 0.268 0.53143 -2.47 1.3e-002
agesite 0.03235 1.033 0.01608 2.01 4.4e-002
racesite 0.85014 2.340 0.24774 3.43 6.0e-004
Rsquare= 0.11 (max possible= 1 )
Likelihood ratio test= 67.1 on 10 df, p=1.58e-010
Wald test = 66.5 on 10 df, p=2.07e-010
Score (logrank) test = 67.4 on 10 df, p=1.38e-010