Version info: Code for this page was tested in R Under development (unstable) (2012-07-05 r59734)
On: 2012-07-08
With: knitr 0.6.3
Please Note: The purpose of this page is to show how to use various data analysis commands. It does not cover all aspects of the research process which researchers are expected to do. In particular, it does not cover data cleaning and checking, verification of assumptions, model diagnostics and potential follow-up analyses.
Examples of random coefficient poisson models
Poisson models are useful for count data. Examples could include number of traffic tickets an individual receives in a year, number of tumor sites in cancer patients, or number of awards received by students. In each of these cases, we might expect most people to have very few, with a relatively small number of individuals having higher numbers.
Background on the Poisson distribution
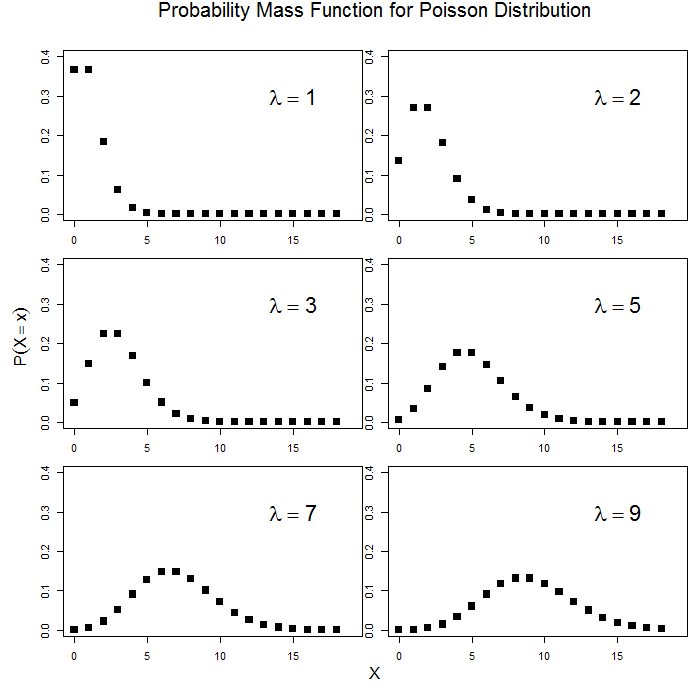
Unlike the familiar Gaussian distribution which has two parameters (mathcal{N}(mu, sigma^{2})), the Poisson distribution is described by a single parameter, (lambda) that is both the mean and variance. That is, (lambda = E(x)) and (lambda = Var(x) = E(x^{2}) – E(x)^{2}). In many ways, this can be a strong assumption. In some cases, use of the negative binomial distribution which estimates a second scale parameter to allow for overdispersion. Here is what the Poisson distribution looks like for different values of (lambda).
par(mfrow = c(3, 2), mar = c(1, 1, 2, 1), oma = c(4, 4, 2, 0)) for (i in c(1, 2, 3, 5, 7, 9)) { curve(dpois(x, lambda = i), from = 0, to = 18, n = 19, type = "p", pch = 15, cex = 1.5, xlim = c(0, 19), ylim = c(0, 0.4), xlab = "", ylab = "") text(x = 15, y = 0.3, substitute(lambda == x, list(x = i)), cex = 2) } mtext("Probability Mass Function for Poisson Distribution", line = 0.5, outer = TRUE, cex = 1.2) mtext(expression(P(X == x)), side = 2, line = 1.5, outer = TRUE) mtext("X", side = 1, line = 1.5, outer = TRUE)
Without random coefficients, the standard Poisson model is:
\begin{equation} \log E(y_{i}) = \alpha + X’_{i} \beta \end{equation}
The log link is the canonical link function for the Poisson distribution, and the expected value of the response is modeled.
With random coefficients, for example a random intercept, the model becomes:
\begin{equation} \log E(y_{ij}|u_{j}) = \alpha + X’_{ij} \beta + u_{j} \end{equation}
Where \(y_{ij}\) is the observation for individual (i) in group (j) and \(u_{j}\) is the random effect for group (j). Thus the two distributions are:
\begin{equation} y \sim Pois(\lambda) \end{equation}
and
\begin{equation} u \sim N(0, \sigma^{2}) \end{equation}
The random coefficient model is conditional on the random effect. To show what this means, consider a simple model:
\begin{equation} \log E(y_{ij}|u_{j}) = -.5 + .3x_{ij} + u_{j} \end{equation}
In the original units, this becomes:
\begin{equation} E(y_{ij}|u_{j}) = \exp(-.5 + .3x_{ij} + u_{j}) \end{equation}
Now look what happens when we graph the estimated change for a 1 unit change in x for values of the random variable (u) ranging from 0 to 4 by increments of .5.
par(mfrow=c(1,1)) f <- function(x_ij, u_j) {
exp(-0.5 + x_ij * 0.3 + u_j) } for (i in seq(0, 4, 0.5)) { curve(f(x, u_j = i), from = 0, to = 1, n = 200, add = i > 0, ylim = c(0, 45), ylab = "") } title(ylab = bquote(E(Y ~ l ~ x[ij], u[j])), main = "Effect of x for different random effects")
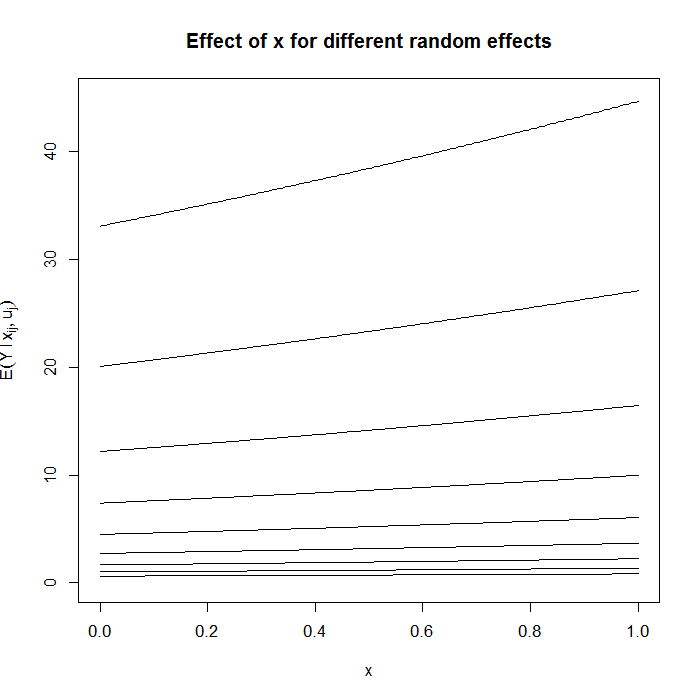
Clearly, on the scale of the original units, a 1 unit increase in x has different effects depending on the value of u, hence the conditionalness of the model. Population “average” effects can be obtained by integrating out the random effect or by fitting a marginal model such as using GEEs.
Although the outcome is assumed to have a Poisson distribution, the random effect (in the above example, u) is typically assumed to have a Gaussian distribution.
Description of the data
The example data we use comes from a sample of the high school and beyond data set, with a made up variable, number of awards a student receives, awards. Our main predictor will be sex, female, and students are clustered (grouped) within schools, cid. First, we will load all the packages required for these examples.
## load foreign package to read Stata data files require(foreign) ## ggplot2 package for graphs require(ggplot2)
You need to install the glmmADMB() package via instructions given in http://glmmadmb.r-forge.r-project.org/. Note that if you are not able to install the glmmADMB() package you may need to use Rtools and compile from source, use Linux, or ask the authors of the package for more information.
install.packages("R2admb")
install.packages("glmmADMB",
repos="http://glmmadmb.r-forge.r-project.org/repos",
type="source")
require(glmmADMB)
## load lme4 package require(lme4)
## read in data dat <- read.dta("https://stats.idre.ucla.edu/stat/data/hsbdemo.dta") dat$cid <- factor(dat$cid) ## look at the first few rows of the dataset head(dat)
id female ses schtyp prog read write math science socst honors awards cid 1 45 female low public vocation 34 35 41 29 26 not enrolled 0 1 2 108 male middle public general 34 33 41 36 36 not enrolled 0 1 3 15 male high public vocation 39 39 44 26 42 not enrolled 0 1 4 67 male low public vocation 37 37 42 33 32 not enrolled 0 1 5 153 male middle public vocation 39 31 40 39 51 not enrolled 0 1 6 51 female high public general 42 36 42 31 39 not enrolled 0 1
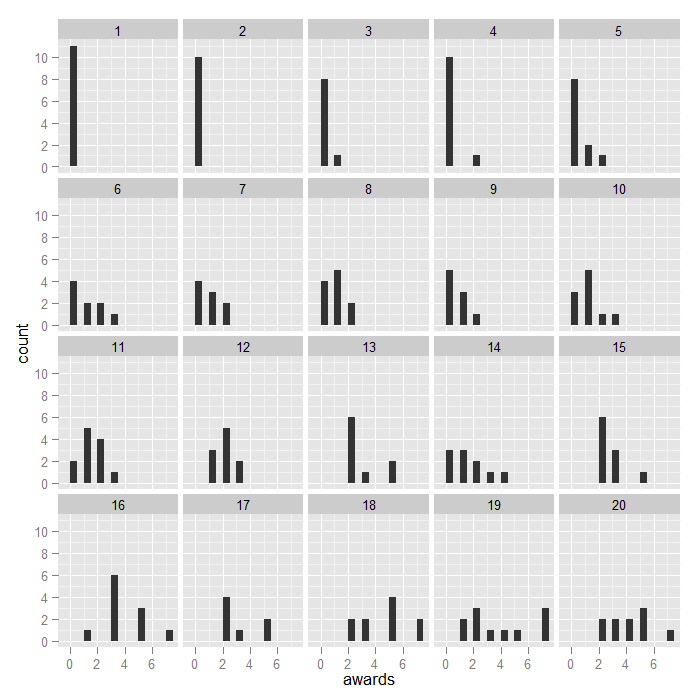
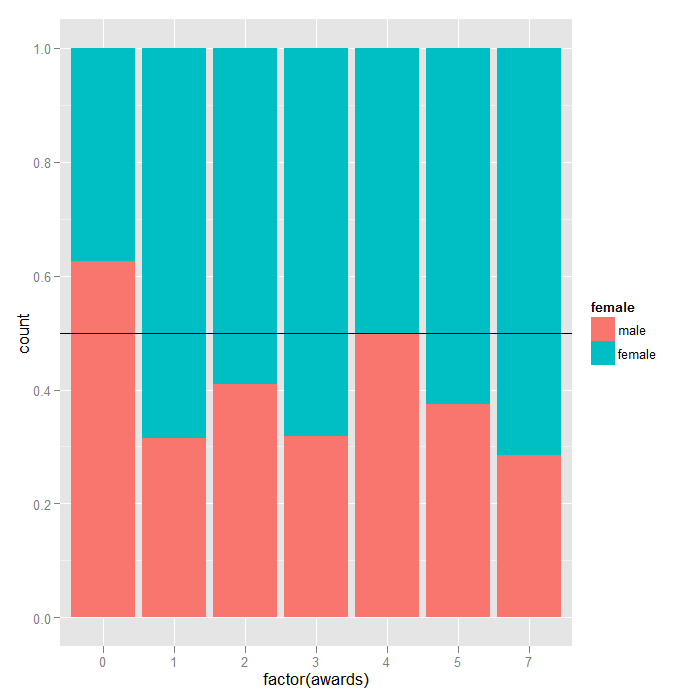
We can get a sense of the distributions in the data using the ggplot2 package. The first plot is just histograms of number of awards for every cid. The second is a filled density plot. The density sums to 1 and the fill shows the distribution of female at every level of awards. If the distribution of female is equal across all awards, they would fall on the horizontal line.
## awards by school
ggplot(dat, aes(awards)) + geom_histogram(binwidth = 0.5) + facet_wrap(~cid)
## density of awards by sex, line at .5 is the null of no sex differences ## in number of awards ggplot(dat, aes(factor(awards))) + geom_bar(aes(fill = female), position = "fill") + geom_hline(yintercept = 0.5)
Analysis methods you might consider
- Random coefficient poisson models, the focus of this page.
- Poisson regression with robust standard errors
Random coefficient poisson model analysis
Because generalized linear mixed models (GLMMs) such as random coefficient poisson models are rather difficult to fit, there tends to be some variability in parameter estimates between different programs. We will demonstrate the use of two packages in R that are able to fit these models, lme4 and glmmADMB.
## fit a random intercept only model using the Laplace approximation ## (equivalent to 1 point evaluated per axis in Gauss-Hermite ## approximation) m1a <- glmer(awards ~ 1 + (1 | cid), data = dat, family = poisson(link = "log")) ## fit a random intercept only model using 100 points per axis in the ## adaptive Gauss-Hermite approximation of the log likelihood more points ## improves accuracy but will take longer m1b <- glmer(awards ~ 1 + (1 | cid), data = dat, family = poisson(link = "log"), nAGQ = 100) ## compare (only slightly different) rbind(m1a = coef(summary(m1a)), m1b = coef(summary(m1b)))
## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.008853 0.2826 -0.03133 0.9750 ## (Intercept) -0.009280 0.2834 -0.03275 0.9739
## view summary output from the more accurate model summary(m1b)
Generalized linear mixed model fit by maximum likelihood (Adaptive Gauss-Hermite Quadrature, nAGQ =
100) [glmerMod]
Family: poisson ( log )
Formula: awards ~ 1 + (1 | cid)
Data: dat
AIC BIC logLik deviance df.resid
228.6 235.2 -112.3 224.6 198
Scaled residuals:
Min 1Q Median 3Q Max
-1.3857 -0.5260 -0.3383 0.3379 3.3769
Random effects:
Groups Name Variance Std.Dev.
cid (Intercept) 1.458 1.207
Number of obs: 200, groups: cid, 20
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.009575 0.290294 -0.033 0.974
We can visually inspect the random effects with a Q-Q plot or a caterpillar plot
## QQ plot plot(ranef(m1b))
## $cid
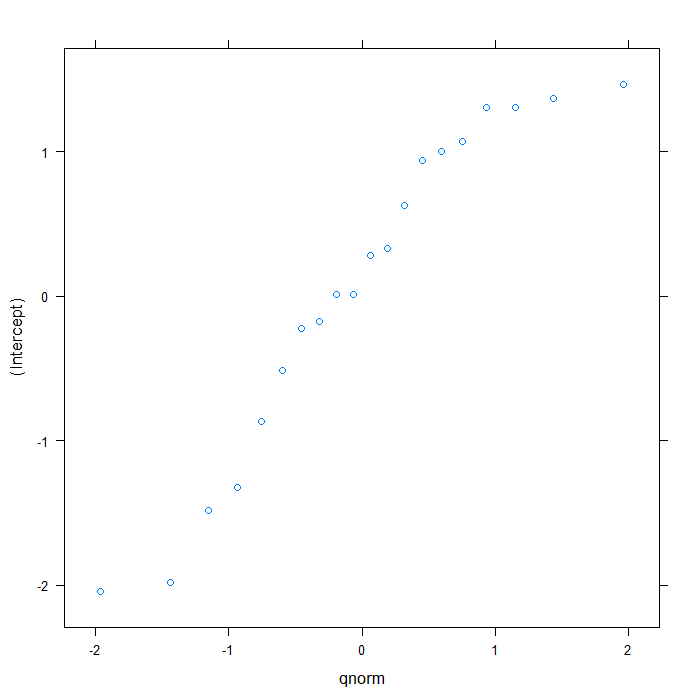
## Caterpillar plot lattice::dotplot(ranef(m1b, postVar = TRUE))
## $cid
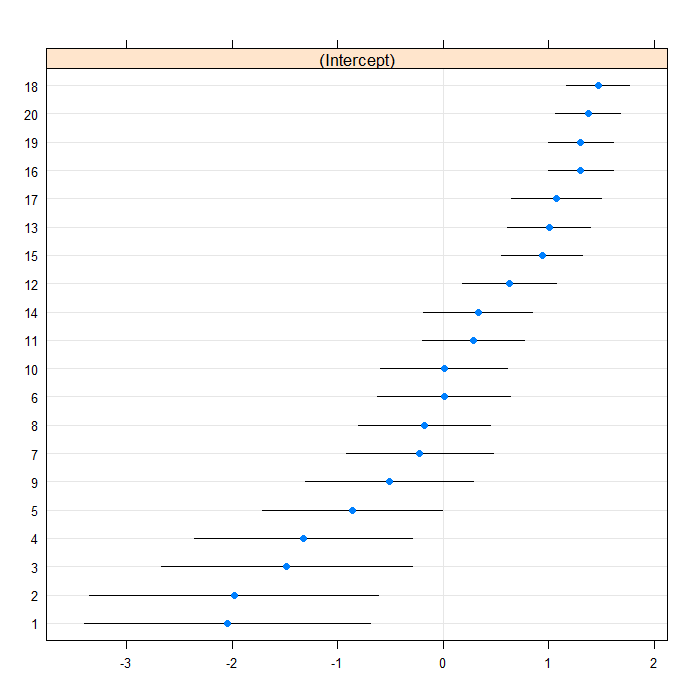
The estimate for the intercept is essentially 0, although the random effects variance indicates that there is some variability in the intercepts between schools. Now we will add in female as an explanatory variable.
m2 <- glmer(awards ~ 1 + female + (1 | cid), data = dat, family = poisson(link = "log"), nAGQ = 100) summary(m2)
Generalized linear mixed model fit by maximum likelihood (Adaptive Gauss-Hermite Quadrature, nAGQ =
100) [glmerMod]
Family: poisson ( log )
Formula: awards ~ 1 + female + (1 | cid)
Data: dat
AIC BIC logLik deviance df.resid
221.1 231.0 -107.6 215.1 197
Scaled residuals:
Min 1Q Median 3Q Max
-1.5312 -0.5919 -0.3304 0.2047 2.8806
Random effects:
Groups Name Variance Std.Dev.
cid (Intercept) 1.431 1.196
Number of obs: 200, groups: cid, 20
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.2229 0.2975 -0.749 0.45370
femalefemale 0.3632 0.1193 3.044 0.00234 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr)
femalefemal -0.252
There appears to be a fairly strong effect of females such that females tend to get more awards than males. Now we will fit the same models using the glmmADMB package
## random intercept only model library(glmmADMB)
m.alt1 <- glmmadmb(awards ~ 1 + (1 | cid), data = dat, family = "poisson", link = "log") m.alt2 <- glmmadmb(awards ~ 1 + female + (1 | cid), data = dat, family = "poisson", link = "log")
summary(m.alt1)
Call:
glmmadmb(formula = awards ~ 1 + (1 | cid), data = dat, family = "poisson",
link = "log")
AIC: 575.1
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.00881 0.28902 -0.03 0.98
Number of observations: total=200, cid=20
Random effect variance(s):
Group=cid
Variance StdDev
(Intercept) 1.443 1.201
Log-likelihood: -285.528
summary(m.alt2)
Call: glmmadmb(formula = awards ~ 1 + female + (1 | cid), data = dat, family = "poisson", link = "log") AIC: 567.5 Coefficients: Estimate Std. Error z value Pr(>|z|) (Intercept) -0.222 0.296 -0.75 0.4534 femalefemale 0.363 0.119 3.04 0.0023 ** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Number of observations: total=200, cid=20 Random effect variance(s): Group=cid Variance StdDev (Intercept) 1.417 1.19 Log-likelihood: -280.769
The results from glmmadmb match closely with those from glmer.
Things to consider
There have been reports (e.g., Zhang et al, 2009) of inconsistency and substantial alpha inflation between different statistical packages and estimation techniques (e.g., using penalized quasi-likelihood, Laplace approximation, Gauss-Hermite quadrature).
See also
Although not the primary focus of this page, because of these known difficulties with GLMMs, we also show the Stata and SAS code to run the same models using a variety of estimation techniques.
SAS
The dataset can be downloaded from here: hsbdemo
data dat; set "c:\temp\hsbdemo.sas7bdat"; run; proc sort data=dat; by cid descending female; run; PROC GLIMMIX data = dat method=LAPLACE noclprint; class cid; model awards = / solution dist=poisson; random intercept / subject=cid; run; PROC GLIMMIX data = dat method=QUAD(QPOINTS = 100) noclprint; class cid; model awards = / solution dist=poisson; random intercept / subject=cid; run; PROC GLIMMIX data = dat method=LAPLACE noclprint order = data; class female cid; model awards = female / solution dist=poisson; random intercept / subject=cid; run; PROC GLIMMIX data = dat method=QUAD(QPOINTS = 100) noclprint order = data; class female cid; model awards = female / solution dist=poisson; random intercept / subject=cid; run; /*We can also manually parameterize the model in NLMIXED */ PROC NLMIXED data = dat METHOD=GAUSS QPOINTS=100; eta = alpha + u; lambda = exp(eta); model awards ~ poisson(lambda); random u ~ normal(0, exp(2*log_sigma)) subject=cid; estimate 'intercept variance' exp(2*log_sigma); run; PROC NLMIXED data = dat METHOD=GAUSS QPOINTS=100; eta = alpha + beta*female + u; lambda = exp(eta); model awards ~ poisson(lambda); random u ~ normal(0, exp(2*log_sigma)) subject=cid; estimate 'intercept variance' exp(2*log_sigma); run;
Random intercept only model, Laplace approximation)
Covariance Parameter Estimates Cov Parm Subject Estimate Standard Error Intercept CID 1.4429 0.5970
Random intercept only model, adaptive Gauss-Hermite approximation with 100 evaluation points)
Random intercept plus female model, adaptive Gauss-Hermite approximation with 100 evaluation points)
Random intercept only model, adaptive Gauss-Hermite approximation with 100 evaluation points) using PROC NLMIXED
| Parameter Estimates | ||||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | Estimate | Standard Error | DF | t Value | Pr > |t| | 95% Confidence Limits | Gradient | |
| alpha | -0.00957 | 0.2903 | 19 | -0.03 | 0.9740 | -0.6172 | 0.5980 | 2.881E-6 |
| log_sigma | 0.1885 | 0.2077 | 19 | 0.91 | 0.3756 | -0.2463 | 0.6233 | 7.13E-8 |
| Additional Estimates | ||||||||
|---|---|---|---|---|---|---|---|---|
| Label | Estimate | Standard Error | DF | t Value | Pr > |t| | Alpha | Lower | Upper |
| intercept variance | 1.4579 | 0.6057 | 19 | 2.41 | 0.0264 | 0.05 | 0.1901 | 2.7257 |
Random intercept plus female model, adaptive Gauss-Hermite approximation with 100 evaluation points) using PROC NLMIXED
Parameter Estimates Parameter Estimate Standard Error DF t Value Pr > |t| 95% Confidence Limits Gradient alpha -0.2229 0.2975 19 -0.75 0.4629 -0.8456 0.3998 8.17E-8 beta 0.3632 0.1193 19 3.04 0.0067 0.1134 0.6129 -0.00004 log_sigma 0.1793 0.2079 19 0.86 0.3991 -0.2558 0.6145 9.151E-6
Additional Estimates Label Estimate Standard Error DF t Value Pr > |t| Alpha Lower Upper intercept variance 1.4314 0.5952 19 2.40 0.0265 0.05 0.1856 2.6773
Stata
use https://stats.idre.ucla.edu/stat/data/hsbdemo, clear *Random intercept model, Laplace xtmepoisson awards || cid:, laplace var *Random intercept model, adaptive Gauss-Hermite, 100 points xtmepoisson awards || cid:, intpoints(100) var *Random intercept and female xtmepoisson awards female || cid:, laplace var *Random intercept and female xtmepoisson awards female || cid:, intpoints(100) var
Random intercept only model, Laplace approximation
------------------------------------------------------------------------------
awards | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
_cons | -.0088 .2890208 -0.03 0.976 -.5752703 .5576703
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
cid: Identity |
var(_cons) | 1.442954 .5970273 .6413056 3.246685
------------------------------------------------------------------------------
Random intercept only model, adaptive Gauss-Hermite approximation with 100 evaluation points)
------------------------------------------------------------------------------
awards | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
_cons | -.0095717 .2902932 -0.03 0.974 -.5785359 .5593924
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
cid: Identity |
var(_cons) | 1.45791 .6057226 .645772 3.291411
------------------------------------------------------------------------------
Random intercept plus female model, Laplace approximation
------------------------------------------------------------------------------
awards | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
female | .3632063 .1193049 3.04 0.002 .129373 .5970396
_cons | -.2221984 .2963088 -0.75 0.453 -.8029531 .3585562
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
cid: Identity |
var(_cons) | 1.416836 .5867228 .6292602 3.190133
------------------------------------------------------------------------------
Random intercept plus female model, adaptive Gauss-Hermite approximation with 100 evaluation points)
------------------------------------------------------------------------------
awards | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
female | .3631578 .1193071 3.04 0.002 .1293202 .5969954
_cons | -.2229269 .29752 -0.75 0.454 -.8060553 .3602015
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
cid: Identity |
var(_cons) | 1.431444 .5952188 .6336221 3.23384
------------------------------------------------------------------------------
References
- Categorical Data Analysis 2nd Ed. by Alan Agresti
- Regression Analysis of Count Data by Cameron and Trivedi
- Zhang, Lu, Feng, Thurston, Xia, Zhu, & Tu, 2009. On fitting generalized linear mixed-effects models for binary responses using different statistical packages. Statistics in Medicine. DOI: 10.1002/sim.4265.
