Version info: Code for this page was tested in SAS 9.3.
Zero-truncated negative binomial regression is used to model count data for which the value zero cannot occur and when there is evidence of over dispersion .
Please Note: The purpose of this page is to show how to use various data analysis commands. It does not cover all aspects of the research process which researchers are expected to do. In particular, it does not cover data cleaning and verification, verification of assumptions, model diagnostics and potential follow-up analyses.
Examples of zero-truncated negative binomial
Example 1.
A study of the length of hospital stay, in days, as a function of age, kind of health insurance and whether or not the patient died while in the hospital. Length of hospital stay is recorded as a minimum of at least one day.
Example 2.
A study of the number of journal articles published by tenured faculty as a function of discipline (fine arts, science, social science, humanities, medical, etc). To get tenure faculty must publish, i.e., there are no tenured faculty with zero publications.
Example 3.
A study by the county traffic court on the number of tickets received by teenagers as predicted by school performance, amount of driver training and gender. Only individuals who have received at least one citation are in the traffic court files.
Description of the data
Let’s pursue Example 1 from above.
We have a hypothetical data file, ztp.sas7bdat with 1,493 observations available here .
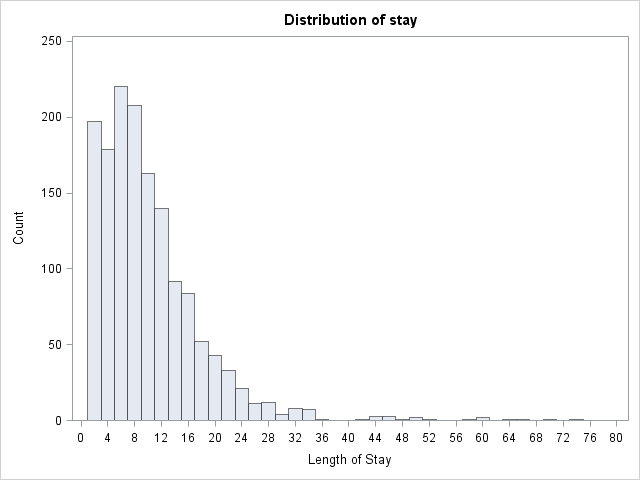
The variable describing length of hospital visit is stay
Let’s look at the data.
proc means data=mylib.ztp;
var stay;
run;
The MEANS Procedure
Analysis Variable : stay Length of Stay
N Mean Std Dev Minimum Maximum
--------------------------------------------------------------------
1493 9.7287341 8.1329081 1.0000000 74.0000000
--------------------------------------------------------------------
proc univariate data=mylib.ztp noprint;
histogram stay / midpoints = 0 to 80 by 2 vscale = count;
run;
 proc freq data=mylib.ztp;
tables age hmo died;
run;
The FREQ Procedure
Age Group
Cumulative Cumulative
age Frequency Percent Frequency Percent
--------------------------------------------------------
1 6 0.40 6 0.40
2 60 4.02 66 4.42
3 163 10.92 229 15.34
4 291 19.49 520 34.83
5 317 21.23 837 56.06
6 327 21.90 1164 77.96
7 190 12.73 1354 90.69
8 93 6.23 1447 96.92
9 46 3.08 1493 100.00
hmo
Cumulative Cumulative
hmo Frequency Percent Frequency Percent
--------------------------------------------------------
0 1254 83.99 1254 83.99
1 239 16.01 1493 100.00
died
Cumulative Cumulative
died Frequency Percent Frequency Percent
---------------------------------------------------------
0 981 65.71 981 65.71
1 512 34.29 1493 100.00
proc freq data=mylib.ztp;
tables age hmo died;
run;
The FREQ Procedure
Age Group
Cumulative Cumulative
age Frequency Percent Frequency Percent
--------------------------------------------------------
1 6 0.40 6 0.40
2 60 4.02 66 4.42
3 163 10.92 229 15.34
4 291 19.49 520 34.83
5 317 21.23 837 56.06
6 327 21.90 1164 77.96
7 190 12.73 1354 90.69
8 93 6.23 1447 96.92
9 46 3.08 1493 100.00
hmo
Cumulative Cumulative
hmo Frequency Percent Frequency Percent
--------------------------------------------------------
0 1254 83.99 1254 83.99
1 239 16.01 1493 100.00
died
Cumulative Cumulative
died Frequency Percent Frequency Percent
---------------------------------------------------------
0 981 65.71 981 65.71
1 512 34.29 1493 100.00
Analysis methods you might consider
Before we show how you can analyze these data with a zero-truncated negative binomial analysis, let’s consider some other methods that you might use.
- Zero-truncated Negative Binomial Regression – The focus of this web page.
- Zero-truncated Poisson Regression – Useful if there is no overdispersion in the zero truncated variable. See the Data Analysis Example for ztp.
- Negative Binomial Regression – Ordinary negative binomial regression will have difficulty with zero-truncated data. It will try to predict zero counts even though there are no zero values.
- Poisson Regression – The same concerns as for negative binomial regression, namely, ordinary poisson regression will have difficulty with zero-truncated data. It will try to predict zero counts even though there are no zero values.
- OLS Regression – You could try to analyze these data using OLS regression. However, count data are highly non-normal and are not well estimated by OLS regression.
Zero-truncated negative binomial regression using proc nlmixed
In order to use proc nlmixed to perform truncated negative binomial regression, we must supply it with a likelihood function. The probability that an observation has count (y) under the negative binomial distribution (without zero truncation) is given by the equation: [P(Y=y) = {y+frac{1}{alpha}-1 choose frac{1}{alpha}-1}left(frac{1}{1+alphamu}right)^{frac{1}{alpha}}left(frac{alphamu}{1+alphamu}right)^y,] where (alpha) is the overdispersion parameter and (mu) is the mean of the negative binomial distribution. With zero truncation, we calculate the probability that (Y=y) conditional on (Y>0), that is, that (Y) is observed as 0 values are not observed. The probability of a zero count under the negative binomial distribution is (P(Y=0) = left(frac{1}{1+alphamu}right)^{frac{1}{alpha}}). The conditional probability is then: [P(Y=y|Y>0) = frac{P(Y=y)}{P(Y>0)} = frac{P(Y=y)}{1-P(Y=0)} = {y+frac{1}{alpha}-1 choose frac{1}{alpha}-1}left(frac{1}{1+alphamu}right)^{frac{1}{alpha}}left(frac{alphamu}{1+alphamu}right)^yfrac{1}{1- left(frac{1}{1+alphamu}right)^{frac{1}{alpha}}}.] The log-likelihood function for the zero-truncated negative binomial distribution is thus: [mathcal{L}=sumlimits_{i=1}^nlogGammaleft(y + frac{1}{alpha}right) – logGammaleft(y+1right) – logGammaleft(frac{1}{alpha}right) – frac{1}{alpha}log(1+alphamu) + ylog(alphamu) – ylog(1 + alphamu) – logleft(1- left(1+alphamuright)^{-frac{1}{alpha}}right).] In negative binomial regression, we model (log(mu)), the log of the mean (expected counts), as a linear combination of a set of predictors: [log(mu) = beta_0 + beta_1x_1 + beta_2x_2 + beta_3x_3] We supply the last two equations to proc nlmixed to model our data using a zero-truncated negative distribution. Additionally, proc nlmixed does not support a class statement, so categorical variables should be dummy-coded before running the analysis. Unlike other SAS procs, by default the first group is the reference group by default in proc nlmixed.
proc nlmixed data = mylib.ztp;
log_mu = intercept + b_age*age + b_died*died + b_hmo*hmo;
mu = exp(log_mu);
het = 1/alpha;
ll = lgamma(stay+het) - lgamma(stay + 1) - lgamma(het) - het*log(1+alpha*mu)
+ stay*log(alpha*mu) - stay*log(1+alpha*mu) - log(1 - (1 + alpha * mu)**-het);
model stay ~ general(ll);
run;
The NLMIXED Procedure
Specifications
Data Set MYLIB.ZTP
Dependent Variable stay
Distribution for Dependent Variable General
Optimization Technique Dual Quasi-Newton
Integration Method None
Dimensions
Observations Used 1493
Observations Not Used 0
Total Observations 1493
Parameters 5
Parameters
intercept b_age b_died b_hmo alpha NegLogLike
1 1 1 1 1 10136.7274
Iteration History
Iter Calls NegLogLike Diff MaxGrad Slope
1 3 5203.75757 4932.97 1718.332 -825332
2 6 5130.65185 73.10572 212.6078 -12208.4
3 8 4922.88698 207.7649 1701.184 -735.733
4 9 4862.95248 59.9345 176.3689 -177.538
5 11 4851.81702 11.13546 393.0774 -13.9647
6 12 4838.27102 13.546 179.7832 -7.96192
7 16 4788.46175 49.80926 168.3697 -26.6674
8 17 4774.94754 13.51421 105.3687 -117.309
9 18 4759.72531 15.22222 77.4436 -25.9074
10 20 4755.95435 3.77096 85.88275 -22.2361
11 22 4755.3438 0.610557 39.18804 -2.65095
12 24 4755.29354 0.050252 30.83521 -0.14278
13 26 4755.28066 0.012889 3.944229 -0.03589
14 28 4755.28014 0.000512 0.44716 -0.00416
15 29 4755.27964 0.0005 0.195745 -0.00109
16 31 4755.27962 0.000028 0.007496 -0.00006
17 33 4755.27962 1.109E-7 0.030916 -4.12E-7
NOTE: GCONV convergence criterion satisfied.
The SAS System 09:40 Monday, June 4, 2012 5
The NLMIXED Procedure
Fit Statistics
-2 Log Likelihood 9510.6
AIC (smaller is better) 9520.6
AICC (smaller is better) 9520.6
BIC (smaller is better) 9547.1
Parameter Estimates
Standard
Parameter Estimate Error DF t Value Pr > |t| Alpha Lower Upper Gradient
intercept 2.4083 0.07198 1493 33.46 <.0001 0.05 2.2671 2.5495 -0.00749
b_age -0.01569 0.01311 1493 -1.20 0.2314 0.05 -0.04140 0.01002 -0.03092
b_died -0.2178 0.04616 1493 -4.72 <.0001 0.05 -0.3083 -0.1272 0.000972
b_hmo -0.1471 0.05922 1493 -2.48 0.0131 0.05 -0.2632 -0.03090 -0.00018
alpha 0.5663 0.03123 1493 18.13 <.0001 0.05 0.5050 0.6276 -0.00461
The output looks very much like the output from an OLS regression:
- Close to the top is the iteration log giving the values of the log likelihoods starting with a model that has no predictors. The last value in the log (4755.27962) is the final value of the negative log likelihood for the full model and is repeated below.
- Next comes a number of fit statistics, which can be used to compare the fit of nested models.
- Below the header you will find the zero-truncated negative binomial coefficients for each of the variables along with standard errors, t-values, p-values and 95% confidence intervals for each coefficient.
- Below that, is the the overdispersion parameter alpha along with its standard error, t-value, p-value, and 95% confidence interval.
Looking through the results we see the following:
- The value of the coefficient for age, -.01569, suggests that the log count of stay decreases by .01569 for each unit increase in age group. This coefficient is not statistically significant.
- The coefficient for hmo, -.1471, is significant and indicates that the log count of stay for HMO patient is .1471 less than for non-HMO patients.
- The log count of stay for patients that died while in the hospital was .2178 less than those patients that did not die.
- The value of the constant (intercept), 2.4083 is log count of the stay when all of the predictors equal zero.
- The estimate for alpha is .5663. For comparison, a model with an alpha of zero is equivalent to a zero-truncated poisson model. In this model, alpha is statistically different from zero, suggesting that the negative binomial model is a better choice than a poisson model.
We can also use estimate statments to help understand our model, by examining the predicted or expected length of stay of patients with varying covariate values. For example we can predict the expected number of days spent at the hospital across age groups for the two hmo statuses for patients who died. The estimate statement for proc nlmixed works slightly differently from how it works within other procs. Here, each parameter must be explicitly multiplied by the value at which is to be held for that estimate statment, and expressions are allowed, such as exponentiation (see code below). Because we would like to predict actual number of days rather than log number of days, we need to exponentiate the estimate. Additionally, the following expected counts are unconditional, meaning these are the expected lengths of stay for patients with the given covariate values in the entire population, not for those patients who we know have stayed at least one day in the hospital (the conditional expectation).
proc nlmixed data = mylib.ztp;
log_mu = intercept + b_age*age + b_died*died + b_hmo*hmo;
mu = exp(log_mu);
het = 1/alpha;
ll = lgamma(stay+het) - lgamma(stay + 1) - lgamma(het) - het*log(1+alpha*mu)
+ stay*log(alpha*mu) - stay*log(1+alpha*mu) - log(1 - (1 + alpha * mu)**-het);
model stay ~ general(ll);
estimate 'age 1 died 1 hmo 0' exp(intercept * 1 + b_age * 1 + b_died * 1 + b_hmo * 0);
estimate 'age 1 died 1 hmo 1' exp(intercept * 1 + b_age * 1 + b_died * 1 + b_hmo * 1);
estimate 'age 3 died 1 hmo 0' exp(intercept * 1 + b_age * 3 + b_died * 1 + b_hmo * 0);
estimate 'age 3 died 1 hmo 1' exp(intercept * 1 + b_age * 3 + b_died * 1 + b_hmo * 1);
estimate 'age 5 died 1 hmo 0' exp(intercept * 1 + b_age * 5 + b_died * 1 + b_hmo * 0);
estimate 'age 5 died 1 hmo 1' exp(intercept * 1 + b_age * 5 + b_died * 1 + b_hmo * 1);
estimate 'age 7 died 1 hmo 0' exp(intercept * 1 + b_age * 7 + b_died * 1 + b_hmo * 0);
estimate 'age 7 died 1 hmo 1' exp(intercept * 1 + b_age * 7 + b_died * 1 + b_hmo * 1);
estimate 'age 9 died 1 hmo 0' exp(intercept * 1 + b_age * 9 + b_died * 1 + b_hmo * 0);
estimate 'age 9 died 1 hmo 1' exp(intercept * 1 + b_age * 9 + b_died * 1 + b_hmo * 1);
run;
<**SOME OUTPUT OMITTED**>
Additional Estimates
Standard
Label Estimate Error DF t Value Pr > |t| Alpha Lower Upper
age 1 died 1 hmo 0 8.8010 0.6291 1493 13.99 <.0001 0.05 7.5668 10.0351
age 1 died 1 hmo 1 7.5974 0.6545 1493 11.61 <.0001 0.05 6.3135 8.8813
age 3 died 1 hmo 0 8.5290 0.4385 1493 19.45 <.0001 0.05 7.6688 9.3893
age 3 died 1 hmo 1 7.3627 0.5212 1493 14.13 <.0001 0.05 6.3404 8.3850
age 5 died 1 hmo 0 8.2655 0.3256 1493 25.39 <.0001 0.05 7.6268 8.9042
age 5 died 1 hmo 1 7.1352 0.4498 1493 15.86 <.0001 0.05 6.2530 8.0174
age 7 died 1 hmo 0 8.0101 0.3430 1493 23.35 <.0001 0.05 7.3373 8.6830
age 7 died 1 hmo 1 6.9147 0.4540 1493 15.23 <.0001 0.05 6.0242 7.8052
age 9 died 1 hmo 0 7.7627 0.4586 1493 16.93 <.0001 0.05 6.8630 8.6623
age 9 died 1 hmo 1 6.7011 0.5200 1493 12.89 <.0001 0.05 5.6811 7.7211
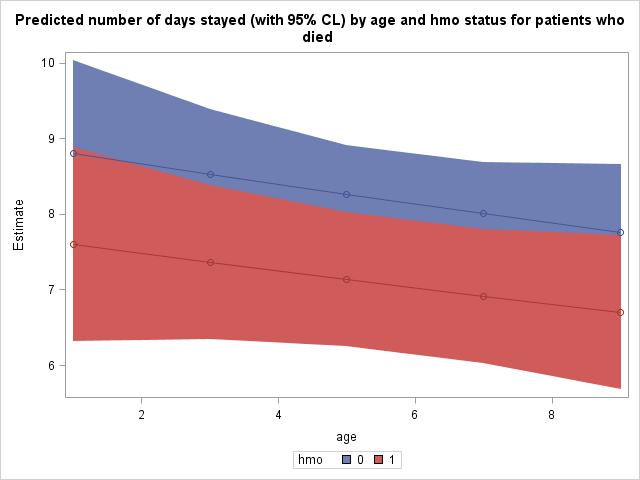
The expected stay for non-HMO patients in age group 1 who died was 8.8010 days, while it was 7.5974 days for HMO patients in age group 1 who died.
We can also test whether the effect of HMO is significant at each level of age for patients who died. We can simply subtract the two estimates that vary by hmo at each level of age.
proc nlmixed data = mylib.ztp;
log_mu = intercept + b_age*age + b_died*died + b_hmo*hmo;
mu = exp(log_mu);
het = 1/alpha;
ll = lgamma(stay+het) - lgamma(stay + 1) - lgamma(het) - het*log(1+alpha*mu)
+ stay*log(alpha*mu) - stay*log(1+alpha*mu) - log(1 - (1 + alpha * mu)**-het);
model stay ~ general(ll);
estimate 'age 1 died 1 hmo 0 vs 1' exp(intercept * 1 + b_age * 1 + b_died * 1 + b_hmo * 0) -
exp(intercept * 1 + b_age * 1 + b_died * 1 + b_hmo * 1);
estimate 'age 3 died 1 hmo 0 vs 1' exp(intercept * 1 + b_age * 3 + b_died * 1 + b_hmo * 0) -
exp(intercept * 1 + b_age * 3 + b_died * 1 + b_hmo * 1);
estimate 'age 5 died 1 hmo 0 vs 1' exp(intercept * 1 + b_age * 5 + b_died * 1 + b_hmo * 0) -
exp(intercept * 1 + b_age * 5 + b_died * 1 + b_hmo * 1);
estimate 'age 7 died 1 hmo 0 vs 1' exp(intercept * 1 + b_age * 7 + b_died * 1 + b_hmo * 0) -
exp(intercept * 1 + b_age * 7 + b_died * 1 + b_hmo * 1);
estimate 'age 9 died 1 hmo 0 vs 1' exp(intercept * 1 + b_age * 9 + b_died * 1 + b_hmo * 0) -
exp(intercept * 1 + b_age * 9 + b_died * 1 + b_hmo * 1);
run;
<**SOME OUTPUT OMITTED**>
Additional Estimates
Standard
Label Estimate Error DF t Value Pr > |t| Alpha Lower Upper
age 1 died 1 hmo 0 vs 1 1.2036 0.4698 1493 2.56 0.0105 0.05 0.2820 2.1251
age 3 died 1 hmo 0 vs 1 1.1664 0.4511 1493 2.59 0.0098 0.05 0.2816 2.0512
age 5 died 1 hmo 0 vs 1 1.1303 0.4350 1493 2.60 0.0095 0.05 0.2770 1.9837
age 7 died 1 hmo 0 vs 1 1.0954 0.4215 1493 2.60 0.0094 0.05 0.2686 1.9222
age 9 died 1 hmo 0 vs 1 1.0616 0.4103 1493 2.59 0.0098 0.05 0.2568 1.8664
The effect of hmo is significant for each age group tested.
It may be illustrative for us to plot the predicted number of days stayed as a function of age and hmo status. To do this, we must tell SAS to save this table of predicted values as a dataset. Tables and graphics produced by procedures are given names upon creation. We will need the name of this prediction table to tell SAS to save it. Place ods trace on and ods trace off statements around the procedure which produced this table to obtain its name. Output from the ods trace statements is located in the log, not the output.
ods trace on;
proc nlmixed data = mylib.ztp;
log_mu = intercept + b_age*age + b_died*died + b_hmo*hmo;
mu = exp(log_mu);
het = 1/alpha;
ll = lgamma(stay+het) - lgamma(stay + 1) - lgamma(het) - het*log(1+alpha*mu)
+ stay*log(alpha*mu) - stay*log(1+alpha*mu) - log(1 - (1 + alpha * mu)**-het);
model stay ~ general(ll);
estimate 'age 1 died 1 hmo 0' exp(intercept * 1 + b_age * 1 + b_died * 1 + b_hmo * 0);
estimate 'age 1 died 1 hmo 1' exp(intercept * 1 + b_age * 1 + b_died * 1 + b_hmo * 1);
estimate 'age 3 died 1 hmo 0' exp(intercept * 1 + b_age * 3 + b_died * 1 + b_hmo * 0);
estimate 'age 3 died 1 hmo 1' exp(intercept * 1 + b_age * 3 + b_died * 1 + b_hmo * 1);
estimate 'age 5 died 1 hmo 0' exp(intercept * 1 + b_age * 5 + b_died * 1 + b_hmo * 0);
estimate 'age 5 died 1 hmo 1' exp(intercept * 1 + b_age * 5 + b_died * 1 + b_hmo * 1);
estimate 'age 7 died 1 hmo 0' exp(intercept * 1 + b_age * 7 + b_died * 1 + b_hmo * 0);
estimate 'age 7 died 1 hmo 1' exp(intercept * 1 + b_age * 7 + b_died * 1 + b_hmo * 1);
estimate 'age 9 died 1 hmo 0' exp(intercept * 1 + b_age * 9 + b_died * 1 + b_hmo * 0);
estimate 'age 9 died 1 hmo 1' exp(intercept * 1 + b_age * 9 + b_died * 1 + b_hmo * 1);
run;
ods trace off;
<***SOME OF THE LOG OMITTED***>
Output Added:
-------------
Name: AdditionalEstimates
Label: Additional Estimates
Template: Stat.NLM.AdditionalEstimates
Path: Nlmixed.AdditionalEstimates
-------------
NOTE: PROCEDURE NLMIXED used (Total process time):
real time 0.23 seconds
cpu time 0.17 seconds
105 ods trace off;
Towards the end of the log we find the name of this table, which as expected by its heading in the output above, is "AdditionalEstimates". We can now tell SAS to save this output table as the dataset "mylib.addest" using an ods output statement.
ods output AdditionalEstimates = mylib.addest;
proc nlmixed data = mylib.ztp;
log_mu = intercept + b_age*age + b_died*died + b_hmo*hmo;
mu = exp(log_mu);
het = 1/alpha;
ll = lgamma(stay+het) - lgamma(stay + 1) - lgamma(het) - het*log(1+alpha*mu)
+ stay*log(alpha*mu) - stay*log(1+alpha*mu) - log(1 - (1 + alpha * mu)**-het);
model stay ~ general(ll);
estimate 'age 1 died 1 hmo 0' exp(intercept * 1 + b_age * 1 + b_died * 1 + b_hmo * 0);
estimate 'age 1 died 1 hmo 1' exp(intercept * 1 + b_age * 1 + b_died * 1 + b_hmo * 1);
estimate 'age 3 died 1 hmo 0' exp(intercept * 1 + b_age * 3 + b_died * 1 + b_hmo * 0);
estimate 'age 3 died 1 hmo 1' exp(intercept * 1 + b_age * 3 + b_died * 1 + b_hmo * 1);
estimate 'age 5 died 1 hmo 0' exp(intercept * 1 + b_age * 5 + b_died * 1 + b_hmo * 0);
estimate 'age 5 died 1 hmo 1' exp(intercept * 1 + b_age * 5 + b_died * 1 + b_hmo * 1);
estimate 'age 7 died 1 hmo 0' exp(intercept * 1 + b_age * 7 + b_died * 1 + b_hmo * 0);
estimate 'age 7 died 1 hmo 1' exp(intercept * 1 + b_age * 7 + b_died * 1 + b_hmo * 1);
estimate 'age 9 died 1 hmo 0' exp(intercept * 1 + b_age * 9 + b_died * 1 + b_hmo * 0);
estimate 'age 9 died 1 hmo 1' exp(intercept * 1 + b_age * 9 + b_died * 1 + b_hmo * 1);
run;
Now we can use this predicted values for plotting. We need to add actual values of age and hmo to the dataset for plotting as well.
data mylib.addest; set mylib.addest; input age hmo; datalines; 1 0 1 1 3 0 3 1 5 0 5 1 7 0 7 1 9 0 9 1 ; run;
Finally, we use proc sgplot to plot our predicted number of days stayed as well as 95% confidence interval bands. The predicted values, lines connecting them, and confidence interval bands are all specified separately within the same proc sgplot. The group option will produce separate points, lines, and bands by the grouping variable.
proc sgplot data = mylib.addest; title 'Predicted number of days stayed (with 95% CL) by age and hmo status for patients who died'; band x = age lower = lower upper = upper / group=hmo; scatter x= age y = estimate / group = hmo; series x = age y = estimate / group = hmo; run;
Things to consider
- Count data often use exposure variable to indicate the number of times the event could have happened. You can incorporate exposure into your model by including a log-linear term for exposure in the log-likehood function specification.
- It is not recommended that zero-truncated negative binomial models be applied to small samples. What constitutes a small sample does not seem to be clearly defined in the literature.
References
- Cameron, A. Colin and Trivedi, P.K. (2009). Microeconometrics using stata. College Station, TX: Stata Press.
- Cameron, A. Colin and Trivedi, P.K. (1998). Regression analysis of count data. Cambridge, UK: Cambridge University Press.
- Hilbe, J. M. (2007). Negative binomial regression. Cambridge, UK: Cambridge University Press.