5.2.2 The Hosmer-Lemeshow Tests
page 150 Table 5.1 Observed (obs) and estimated expected (exp) frequencies within each decile of risk, defined by fitted value (prob.) for dfree = 1 and dfree = 0 using the fitted logistic regression model in Table 4.9.
NOTE: Pursuant to the text on page 151 this table cannot be replicated in SAS. You can use Stata to obtain these values.
5.2.3 Classification tables
page 157 Table 5.2 Classification table based on the logistic regression model in Table 4.9 using a cutpoint of 0.5.
NOTE: We have bolded the relevant output.
data uis51;
set 'd:hosmerdatauis';
ndrgfp1 = ((ndrugtx+1)/10)**(-1);
ndrgfp2 = ndrgfp1*log((ndrugtx+1)/10);
agendrgfp1 = age*ndrgfp1;
racesite = race*site;
run;
proc logistic data=uis51 desc;
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
/ details ctable lackfit pprob = .5;
run;
The LOGISTIC Procedure
Model Information
Data Set WORK.UIS51
Response Variable DFREE
Number of Response Levels 2
Number of Observations 575
Link Function Logit
Optimization Technique Fisher's scoring
Response Profile
Ordered Total
Value DFREE Frequency
1 1 147
2 0 428
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept
Intercept and
Criterion Only Covariates
AIC 655.729 619.963
SC 660.083 667.861
-2 Log L 653.729 597.963
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 55.7660 10 <.0001
Score 52.0723 10 <.0001
Wald 47.2784 10 <.0001
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001
AGE 1 0.1166 0.0289 16.3137 <.0001
ndrgfp1 1 1.6687 0.4071 16.8000 <.0001
ndrgfp2 1 0.4336 0.1169 13.7585 0.0002
IVHX2 1 -0.6346 0.2987 4.5134 0.0336
IVHX3 1 -0.7049 0.2616 7.2623 0.0070
RACE 1 0.6841 0.2641 6.7074 0.0096
TREAT 1 0.4349 0.2038 4.5559 0.0328
SITE 1 0.5162 0.2549 4.1013 0.0429
agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113
racesite 1 -1.4294 0.5298 7.2799 0.0070
Odds Ratio Estimates
Point 95% Wald
Effect Estimate Confidence Limits
AGE 1.124 1.062 1.189
ndrgfp1 5.306 2.389 11.784
ndrgfp2 1.543 1.227 1.940
IVHX2 0.530 0.295 0.952
IVHX3 0.494 0.296 0.825
RACE 1.982 1.181 3.326
TREAT 1.545 1.036 2.303
SITE 1.676 1.017 2.761
agendrgfp1 0.985 0.973 0.997
racesite 0.239 0.085 0.676
Association of Predicted Probabilities and Observed Responses
Percent Concordant 69.7 Somers' D 0.398
Percent Discordant 29.9 Gamma 0.399
Percent Tied 0.4 Tau-a 0.152
Pairs 62916 c 0.699
The LOGISTIC Procedure
Partition for the Hosmer and Lemeshow Test
DFREE = 1 DFREE = 0
Group Total Observed Expected Observed Expected
1 58 4 4.10 54 53.90
2 59 6 6.48 53 52.52
3 59 7 8.78 52 50.22
4 60 12 11.14 48 48.86
5 60 16 13.35 44 46.65
6 58 14 15.14 44 42.86
7 58 19 17.92 39 40.08
8 58 23 20.63 35 37.37
9 58 21 24.79 37 33.21
10 47 25 24.67 22 22.33
Hosmer and Lemeshow Goodness-of-Fit Test
Chi-Square DF Pr > ChiSq
2.8735 8 0.9420
Classification Table
Correct Incorrect Percentages
Prob Non- Non- Sensi- Speci- False False
Level Event Event Event Event Correct tivity ficity POS NEG
0.500 13 414 14 134 74.3 8.8 96.7 51.9 24.5
page 159 Table 5.3 Classification table based on the logistic regression model in Table 4.9 using a cutpoint of 0.5, but all probabilities pi-hat < 0.50 are replaced with pi-hat = 0.05 and all probabilities pi-hat >= 0.50 are replaced with pi-hat = 0.95.
NOTE: We were unable to reproduce this table. page 160 Table 5.4 Classification table based on the logistic regression model in Table 4.9 using a cutpoint of 0.5, but all probabilities pi-hat < 0.50 are replaced with pi-hat = 0.45 and all probabilities pi-hat >= 0.50 are replaced with pi-hat = 0.55.
NOTE: We were unable to reproduce this table.
5.2.4 Area under the ROC curve
page 161 Table 5.5 Classification table based on the logistic regression model in Table 4.9 using a cutpoint of 0.6.
proc logistic data=uis51 desc;
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
/ details lackfit ctable pprob = .6;
run;
The LOGISTIC Procedure
Model Information
Data Set WORK.UIS51
Response Variable DFREE
Number of Response Levels 2
Number of Observations 575
Link Function Logit
Optimization Technique Fisher's scoring
Response Profile
Ordered Total
Value DFREE Frequency
1 1 147
2 0 428
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept
Intercept and
Criterion Only Covariates
AIC 655.729 619.963
SC 660.083 667.861
-2 Log L 653.729 597.963
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 55.7660 10 <.0001
Score 52.0723 10 <.0001
Wald 47.2784 10 <.0001
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001
AGE 1 0.1166 0.0289 16.3137 <.0001
ndrgfp1 1 1.6687 0.4071 16.8000 <.0001
ndrgfp2 1 0.4336 0.1169 13.7585 0.0002
IVHX2 1 -0.6346 0.2987 4.5134 0.0336
IVHX3 1 -0.7049 0.2616 7.2623 0.0070
RACE 1 0.6841 0.2641 6.7074 0.0096
TREAT 1 0.4349 0.2038 4.5559 0.0328
SITE 1 0.5162 0.2549 4.1013 0.0429
agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113
racesite 1 -1.4294 0.5298 7.2799 0.0070
Odds Ratio Estimates
Point 95% Wald
Effect Estimate Confidence Limits
AGE 1.124 1.062 1.189
ndrgfp1 5.306 2.389 11.784
ndrgfp2 1.543 1.227 1.940
IVHX2 0.530 0.295 0.952
IVHX3 0.494 0.296 0.825
RACE 1.982 1.181 3.326
TREAT 1.545 1.036 2.303
SITE 1.676 1.017 2.761
agendrgfp1 0.985 0.973 0.997
racesite 0.239 0.085 0.676
Association of Predicted Probabilities and Observed Responses
Percent Concordant 69.7 Somers' D 0.398
Percent Discordant 29.9 Gamma 0.399
Percent Tied 0.4 Tau-a 0.152
Pairs 62916 c 0.699
The LOGISTIC Procedure
Partition for the Hosmer and Lemeshow Test
DFREE = 1 DFREE = 0
Group Total Observed Expected Observed Expected
1 58 4 4.10 54 53.90
2 59 6 6.48 53 52.52
3 59 7 8.78 52 50.22
4 60 12 11.14 48 48.86
5 60 16 13.35 44 46.65
6 58 14 15.14 44 42.86
7 58 19 17.92 39 40.08
8 58 23 20.63 35 37.37
9 58 21 24.79 37 33.21
10 47 25 24.67 22 22.33
Hosmer and Lemeshow Goodness-of-Fit Test
Chi-Square DF Pr > ChiSq
2.8735 8 0.9420
Classification Table
Correct Incorrect Percentages
Prob Non- Non- Sensi- Speci- False False
Level Event Event Event Event Correct tivity ficity POS NEG
0.600 3 428 0 144 75.0 2.0 100.0 0.0 25.2
page 161 Table 5.6 Summary of sensitivity, specificity, and 1-specificity for classification tables based on the logistic regression model in Table 4.9 using a cutpoint of 0.05 to 0.60 in increments of 0.05.
proc logistic data=uis51 desc;
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
/ ctable pprob = (.05 to .6 by .05);
run;
The LOGISTIC Procedure
Model Information
Data Set WORK.UIS51
Response Variable DFREE
Number of Response Levels 2
Number of Observations 575
Link Function Logit
Optimization Technique Fisher's scoring
Response Profile
Ordered Total
Value DFREE Frequency
1 1 147
2 0 428
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept
Intercept and
Criterion Only Covariates
AIC 655.729 619.963
SC 660.083 667.861
-2 Log L 653.729 597.963
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 55.7660 10 <.0001
Score 52.0723 10 <.0001
Wald 47.2784 10 <.0001
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001
AGE 1 0.1166 0.0289 16.3137 <.0001
ndrgfp1 1 1.6687 0.4071 16.8000 <.0001
ndrgfp2 1 0.4336 0.1169 13.7585 0.0002
IVHX2 1 -0.6346 0.2987 4.5134 0.0336
IVHX3 1 -0.7049 0.2616 7.2623 0.0070
RACE 1 0.6841 0.2641 6.7074 0.0096
TREAT 1 0.4349 0.2038 4.5559 0.0328
SITE 1 0.5162 0.2549 4.1013 0.0429
agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113
racesite 1 -1.4294 0.5298 7.2799 0.0070
Odds Ratio Estimates
Point 95% Wald
Effect Estimate Confidence Limits
AGE 1.124 1.062 1.189
ndrgfp1 5.306 2.389 11.784
ndrgfp2 1.543 1.227 1.940
IVHX2 0.530 0.295 0.952
IVHX3 0.494 0.296 0.825
RACE 1.982 1.181 3.326
TREAT 1.545 1.036 2.303
SITE 1.676 1.017 2.761
agendrgfp1 0.985 0.973 0.997
racesite 0.239 0.085 0.676
Association of Predicted Probabilities and Observed Responses
Percent Concordant 69.7 Somers' D 0.398
Percent Discordant 29.9 Gamma 0.399
Percent Tied 0.4 Tau-a 0.152
Pairs 62916 c 0.699
Classification Table
Correct Incorrect Percentages
Prob Non- Non- Sensi- Speci- False False
Level Event Event Event Event Correct tivity ficity POS NEG
0.050 146 10 418 1 27.1 99.3 2.3 74.1 9.1
0.100 140 65 363 7 35.7 95.2 15.2 72.2 9.7
0.150 130 134 294 17 45.9 88.4 31.3 69.3 11.3
The LOGISTIC Procedure
Classification Table
Correct Incorrect Percentages
Prob Non- Non- Sensi- Speci- False False
Level Event Event Event Event Correct tivity ficity POS NEG
0.200 113 191 237 34 52.9 76.9 44.6 67.7 15.1
0.250 95 255 173 52 60.9 64.6 59.6 64.6 16.9
0.300 77 302 126 70 65.9 52.4 70.6 62.1 18.8
0.350 53 343 85 94 68.9 36.1 80.1 61.6 21.5
0.400 34 370 58 113 70.3 23.1 86.4 63.0 23.4
0.450 27 391 37 120 72.7 18.4 91.4 57.8 23.5
0.500 13 414 14 134 74.3 8.8 96.7 51.9 24.5
0.550 7 425 3 140 75.1 4.8 99.3 30.0 24.8
0.600 3 428 0 144 75.0 2.0 100.0 0.0 25.2
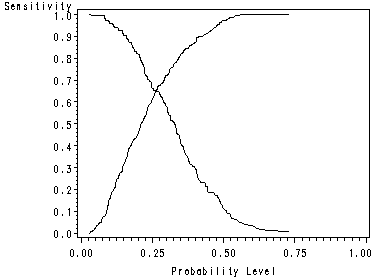
page 162 Figure 5.1 Plot of sensitivity and specificity versus all possible cutpoints in the UIS.
proc logistic data=uis51 desc;
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
/ outroc=roc1;
run;
data roc2;
set roc1;
spec = 1-_1mspec_;
run;
symbol1 i=join v=none ;
proc gplot data=roc2;
plot _sensit_*_PROB_=1 spec*_PROB_=1 / overlay haxis=0 to 1 by .25 vaxis=0 to 1 by .1 ;
run;
quit;
The LOGISTIC Procedure
Model Information
Data Set WORK.UIS51
Response Variable DFREE
Number of Response Levels 2
Number of Observations 575
Link Function Logit
Optimization Technique Fisher's scoring
Response Profile
Ordered Total
Value DFREE Frequency
1 1 147
2 0 428
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept
Intercept and
Criterion Only Covariates
AIC 655.729 619.963
SC 660.083 667.861
-2 Log L 653.729 597.963
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 55.7660 10 <.0001
Score 52.0723 10 <.0001
Wald 47.2784 10 <.0001
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001
AGE 1 0.1166 0.0289 16.3137 <.0001
ndrgfp1 1 1.6687 0.4071 16.8000 <.0001
ndrgfp2 1 0.4336 0.1169 13.7585 0.0002
IVHX2 1 -0.6346 0.2987 4.5134 0.0336
IVHX3 1 -0.7049 0.2616 7.2623 0.0070
RACE 1 0.6841 0.2641 6.7074 0.0096
TREAT 1 0.4349 0.2038 4.5559 0.0328
SITE 1 0.5162 0.2549 4.1013 0.0429
agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113
racesite 1 -1.4294 0.5298 7.2799 0.0070
Odds Ratio Estimates
Point 95% Wald
Effect Estimate Confidence Limits
AGE 1.124 1.062 1.189
ndrgfp1 5.306 2.389 11.784
ndrgfp2 1.543 1.227 1.940
IVHX2 0.530 0.295 0.952
IVHX3 0.494 0.296 0.825
RACE 1.982 1.181 3.326
TREAT 1.545 1.036 2.303
SITE 1.676 1.017 2.761
agendrgfp1 0.985 0.973 0.997
racesite 0.239 0.085 0.676
Association of Predicted Probabilities and Observed Responses
Percent Concordant 69.7 Somers' D 0.398
Percent Discordant 29.9 Gamma 0.399
Percent Tied 0.4 Tau-a 0.152
Pairs 62916 c 0.699
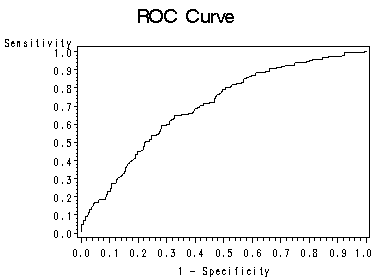
page 163 Figure 5.2 Plot of sensitivity versus 1-specificity for all possible cutpoints in the UIS. The resulting curve is called a ROC curve.
symbol1 i=join v=none ; proc gplot data=roc1; title 'ROC Curve'; plot _sensit_*_1mspec_=1 / vaxis=0 to 1 by .1 ; run; quit; title;
5.3 Logistic regression diagnostics
page 171 Figure 5.3 Plot of leverage (h) versus the estimated logistic probability (pi-hat) for a hypothetical univariable logistic regression model.
NOTE: We cannot recreate this figure because we do have the hypothetical data that were used.
page 172 Figure 5.4 Plot of the distance portion of leverage (b) versus the estimated logistic probability (pi-hat) for a hypothetical univariable logistic regression model.
NOTE: We cannot recreate this figure because we do have the hypothetical data that were used.
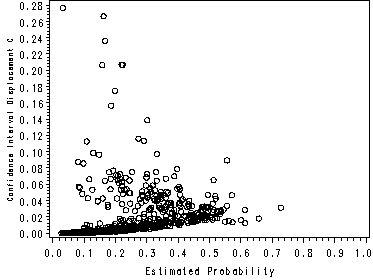
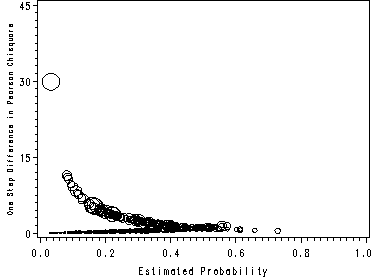
page 177 Figure 5.5 Plot of delta-x-square versus the estimated probability from the fitted model in Table 4.9, UIS J = 521 covariate patterns.
NOTE: This graph looks slightly different than the one in the book because SAS and Stata use different methods of handling covariate patterns. The graphs in the text were made using Stata.
proc logistic data=uis51 desc;
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite / aggregate scale = 1;
output out=uis52 p=estprob DIFCHISQ=deltachi DIFDEV=deltad c=deltabeta;
run;
symbol1 i=none v=circle ;
axis1 order=(0 to 1 by .1) ;
axis2 label=(angle=90 color=black height=0.75);
proc gplot data=uis52;
plot deltachi*estprob=1 / haxis=axis1 vaxis=axis2 ;
run;
quit;
The LOGISTIC Procedure
Model Information
Data Set WORK.UIS51
Response Variable DFREE
Number of Response Levels 2
Number of Observations 575
Link Function Logit
Optimization Technique Fisher's scoring
Response Profile
Ordered Total
Value DFREE Frequency
1 1 147
2 0 428
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Deviance and Pearson Goodness-of-Fit Statistics
Criterion DF Value Value/DF Pr > ChiSq
Deviance 510 530.7412 1.0407 0.2541
Pearson 510 511.7467 1.0034 0.4699
Number of unique profiles: 521
NOTE: The covariance matrix has been multiplied by the heterogeneity factor (square of SCALE=1)
1.
Model Fit Statistics
Intercept
Intercept and
Criterion Only Covariates
AIC 655.729 619.963
SC 660.083 667.861
-2 Log L 653.729 597.963
The LOGISTIC Procedure
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 55.7660 10 <.0001
Score 52.0723 10 <.0001
Wald 47.2784 10 <.0001
Analysis of Maximum Likelihood Estimates
Standard
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001
AGE 1 0.1166 0.0289 16.3137 <.0001
ndrgfp1 1 1.6687 0.4071 16.8000 <.0001
ndrgfp2 1 0.4336 0.1169 13.7585 0.0002
IVHX2 1 -0.6346 0.2987 4.5134 0.0336
IVHX3 1 -0.7049 0.2616 7.2623 0.0070
RACE 1 0.6841 0.2641 6.7074 0.0096
TREAT 1 0.4349 0.2038 4.5559 0.0328
SITE 1 0.5162 0.2549 4.1013 0.0429
agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113
racesite 1 -1.4294 0.5298 7.2799 0.0070
Odds Ratio Estimates
Point 95% Wald
Effect Estimate Confidence Limits
AGE 1.124 1.062 1.189
ndrgfp1 5.306 2.389 11.784
ndrgfp2 1.543 1.227 1.940
IVHX2 0.530 0.295 0.952
IVHX3 0.494 0.296 0.825
RACE 1.982 1.181 3.326
TREAT 1.545 1.036 2.303
SITE 1.676 1.017 2.761
agendrgfp1 0.985 0.973 0.997
racesite 0.239 0.085 0.676
Association of Predicted Probabilities and Observed Responses
Percent Concordant 69.7 Somers' D 0.398
Percent Discordant 29.9 Gamma 0.399
Percent Tied 0.4 Tau-a 0.152
Pairs 62916 c 0.699
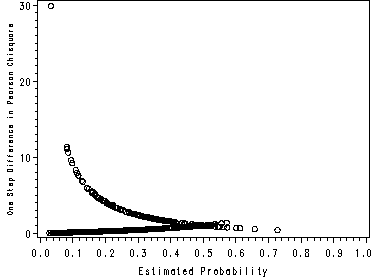
page 178 Figure 5.6 Plot of delta-D versus the estimated probability from the fitted model in Table 4.9, UIS J = 521 covariate patterns.
NOTE: This graph looks slightly different than the one in the book because SAS and Stata use different methods of handling covariate patterns. The graphs in the text were made using Stata.
symbol1 i=none v=circle ; axis1 order=(0 to 1 by .1) ; axis2 label=(angle=90 color=black height=0.75); proc gplot data=uis52; plot deltad*estprob=1 / haxis=axis1 vaxis=axis2; run; quit;
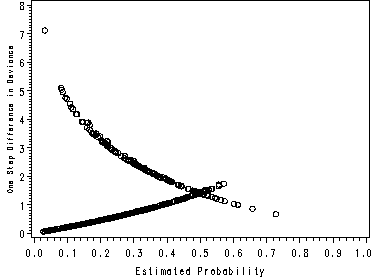
page 179 Figure 5.7 Plot of delta-beta-hat versus the estimated probability from the fitted model in Table 4.9, UIS J = 521 covariate patterns.
NOTE: This graph looks slightly different than the one in the book because SAS and Stata use different methods of handling covariate patterns. The graphs in the text were made using Stata.
symbol1 i=none v=circle ; axis1 order=(0 to 1 by .1) ; axis2 label=(angle=90 color=black height=0.75); proc gplot data=uis52; plot deltabeta*estprob=1 / haxis=axis1 vaxis=axis2; run; quit;
page 180 Figure 5.8 Plot of delta-chi-square versus the probability from the fitted model in Table 4.9 with size of the plotting symbol proportional to delta-beta-hat, UIS J = 521 covariate patterns.
NOTE: This graph looks slightly different than the one in the book because SAS and Stata use different methods of handling covariate patterns. The graphs in the text were made using Stata.
filename outgraph 'd:https://stats.idre.ucla.edu/wp-content/uploads/2016/02/hlch5sas6.gif'; goptions gsfname=outgraph dev=gif373; symbol1 color=black interpol=r value=circle height=1; axis1 order=(0 to 45 by 15) label=(angle=90 color=black height=0.75); axis2 order=(0 to 1 by .2); proc gplot data=uis52; bubble deltachi*estprob=deltabeta / bsize=20 haxis=axis2 vaxis=axis1; run; quit;
page 182 Table 5.8 Covariate values, observed outcome (yj), number (mj), estimated logistic probability (pi-hat), and the value of the four diagnostic statistics delta-beta-hat, delta-x-square, and leverage (h) for the four most extreme covariate patterns (P#).
NOTE: The following must be done to reproduce the covariate patterns as shown in the text, because SAS and Stata handle covariate patterns differently.
proc sort data=uis51 out=uis51sort nodupkey; by age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite; run; proc sort data=uis51 out=uis51sorta; by age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite; run; data uis53; set uis51sort; covpat=_n_; run; data uis54; merge uis51sorta uis53; by age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite; run; proc logistic data=uis54 desc; model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite / aggregate scale = 1; output out=uis55 p=estprob DIFCHISQ=deltachi DIFDEV=deltad c=deltabeta h=lev; run; proc print data=uis55 noobs; var covpat age ndrugtx ivhx race treat site dfree estprob deltabeta deltachi deltad lev; where covpat=31 or covpat=477 or covpat=105 or covpat=468; run;The LOGISTIC Procedure
Model Information
Data Set WORK.UIS54 Response Variable DFREE Number of Response Levels 2 Number of Observations 575 Link Function Logit Optimization Technique Fisher's scoring
Response Profile
Ordered Total Value DFREE Frequency
1 1 147 2 0 428
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Deviance and Pearson Goodness-of-Fit Statistics
Criterion DF Value Value/DF Pr > ChiSq
Deviance 510 530.7412 1.0407 0.2541 Pearson 510 511.7467 1.0034 0.4699
Number of unique profiles: 521
NOTE: The covariance matrix has been multiplied by the heterogeneity factor (square of SCALE=1) 1.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 655.729 619.963 SC 660.083 667.861 -2 Log L 653.729 597.963
The LOGISTIC Procedure
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 55.7660 10 <.0001 Score 52.0723 10 <.0001 Wald 47.2784 10 <.0001
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001 AGE 1 0.1166 0.0289 16.3137 <.0001 ndrgfp1 1 1.6687 0.4071 16.8000 <.0001 ndrgfp2 1 0.4336 0.1169 13.7585 0.0002 IVHX2 1 -0.6346 0.2987 4.5134 0.0336 IVHX3 1 -0.7049 0.2616 7.2623 0.0070 RACE 1 0.6841 0.2641 6.7074 0.0096 TREAT 1 0.4349 0.2038 4.5559 0.0328 SITE 1 0.5162 0.2549 4.1013 0.0429 agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113 racesite 1 -1.4294 0.5298 7.2799 0.0070
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
AGE 1.124 1.062 1.189 ndrgfp1 5.306 2.389 11.784 ndrgfp2 1.543 1.227 1.940 IVHX2 0.530 0.295 0.952 IVHX3 0.494 0.296 0.825 RACE 1.982 1.181 3.326 TREAT 1.545 1.036 2.303 SITE 1.676 1.017 2.761 agendrgfp1 0.985 0.973 0.997 racesite 0.239 0.085 0.676
Association of Predicted Probabilities and Observed Responses
Percent Concordant 69.7 Somers' D 0.398 Percent Discordant 29.9 Gamma 0.399 Percent Tied 0.4 Tau-a 0.152 Pairs 62916 c 0.699
covpat AGE NDRUGTX IVHX RACE TREAT SITE DFREE estprob deltabeta deltachi deltad lev 31 24 20 2 0 0 1 1 0.03264 0.27680 29.9127 7.11877 0.009169 105 26 0 1 1 0 0 1 0.40299 0.05708 1.5365 1.87270 0.035818 105 26 0 1 1 0 0 1 0.40299 0.05708 1.5365 1.87270 0.035818 468 40 0 3 1 0 0 1 0.16762 0.23637 5.1920 3.79822 0.043544 477 41 0 3 1 0 0 1 0.16265 0.26660 5.4023 3.88639 0.047028
page 183 Table 5.9 Estimated coefficients from all data, the percent change when the covariate pattern is deleted, and values of goodness-of-fit statistics for each model.
NOTE: The Hosmer and Lemeshow goodness-of-fit statistic is different than that shown in the text because of the differences in the way SAS and Stata handle ties.
*Column 1 of Table 5.9;
proc logistic data=uis54 desc;
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site
agendrgfp1 racesite / aggregate lackfit scale = 1;
run;
[output omitted]
Deviance and Pearson Goodness-of-Fit Statistics
Criterion Value DF Value/DF Pr > ChiSq
Deviance 530.7412 510 1.0407 0.2541
Pearson 511.7467 510 1.0034 0.4699
Number of unique profiles: 521
[output omitted]
Analysis of Maximum Likelihood Estimates
Standard Wald
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001
AGE 1 0.1166 0.0289 16.3137 <.0001
ndrgfp1 1 1.6687 0.4071 16.8000 <.0001
ndrgfp2 1 0.4336 0.1169 13.7585 0.0002
ivhx2 1 -0.6346 0.2987 4.5134 0.0336
ivhx3 1 -0.7049 0.2616 7.2623 0.0070
RACE 1 0.6841 0.2641 6.7074 0.0096
TREAT 1 0.4349 0.2038 4.5559 0.0328
SITE 1 0.5162 0.2549 4.1013 0.0429
agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113
racesite 1 -1.4294 0.5298 7.2799 0.0070
[output omitted]
Hosmer and Lemeshow Goodness-of-Fit Test
Chi-Square DF Pr > ChiSq
4.4189 8 0.8175
*Column 2 of Table 5.9;
proc logistic data=uis54 desc;
where covpat not in (31);
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site
agendrgfp1 racesite / aggregate lackfit scale = 1;
run;
[output omitted]
Deviance and Pearson Goodness-of-Fit Statistics
Criterion Value DF Value/DF Pr > ChiSq
Deviance 523.6164 509 1.0287 0.3175
Pearson 489.8994 509 0.9625 0.7208
Number of unique profiles: 520
[output omitted]
Analysis of Maximum Likelihood Estimates
Standard Wald
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -7.3714 1.2531 34.6013 <.0001
AGE 1 0.1269 0.0295 18.5683 <.0001
ndrgfp1 1 1.8295 0.4173 19.2219 <.0001
ndrgfp2 1 0.4745 0.1191 15.8807 <.0001
ivhx2 1 -0.6904 0.3028 5.1978 0.0226
ivhx3 1 -0.7087 0.2630 7.2634 0.0070
RACE 1 0.6927 0.2656 6.7993 0.0091
TREAT 1 0.4574 0.2053 4.9651 0.0259
SITE 1 0.4873 0.2572 3.5901 0.0581
agendrgfp1 1 -0.0168 0.00613 7.4808 0.0062
racesite 1 -1.4220 0.5322 7.1385 0.0075
[output omitted]
Hosmer and Lemeshow Goodness-of-Fit Test
Chi-Square DF Pr > ChiSq
4.7204 8 0.7870
*Column 3 of Table 5.9;
proc logistic data=uis54 desc;
where covpat not in (477);
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site
agendrgfp1 racesite / aggregate lackfit scale = 1;
run;
[output omitted]
Deviance and Pearson Goodness-of-Fit Statistics
Criterion Value DF Value/DF Pr > ChiSq
Deviance 526.8477 509 1.0351 0.2830
Pearson 511.5248 509 1.0050 0.4602
Number of unique profiles: 520
[output omitted]
Analysis of Maximum Likelihood Estimates
Standard Wald
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -7.0695 1.2399 32.5072 <.0001
AGE 1 0.1228 0.0294 17.5087 <.0001
ndrgfp1 1 1.7746 0.4183 17.9954 <.0001
ndrgfp2 1 0.4513 0.1183 14.5416 0.0001
ivhx2 1 -0.6375 0.2990 4.5443 0.0330
ivhx3 1 -0.7445 0.2636 7.9757 0.0047
RACE 1 0.6441 0.2660 5.8619 0.0155
TREAT 1 0.4504 0.2045 4.8485 0.0277
SITE 1 0.5162 0.2553 4.0884 0.0432
agendrgfp1 1 -0.0175 0.00629 7.7352 0.0054
racesite 1 -1.3774 0.5319 6.7059 0.0096
[output omitted]
Hosmer and Lemeshow Goodness-of-Fit Test
Chi-Square DF Pr > ChiSq
9.2002 8 0.3257
*Column 4 of Table 5.9;
proc logistic data=uis54 desc;
where covpat not in (105);
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site
agendrgfp1 racesite / aggregate lackfit scale = 1;
run;
[output omitted]
Deviance and Pearson Goodness-of-Fit Statistics
Criterion Value DF Value/DF Pr > ChiSq
Deviance 526.8757 509 1.0351 0.2828
Pearson 508.6675 509 0.9993 0.4958
Number of unique profiles: 520
[output omitted]
Analysis of Maximum Likelihood Estimates
Standard Wald
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.7557 1.2165 30.8427 <.0001
AGE 1 0.1134 0.0289 15.4237 <.0001
ndrgfp1 1 1.6427 0.4065 16.3301 <.0001
ndrgfp2 1 0.4427 0.1170 14.3077 0.0002
ivhx2 1 -0.6368 0.2992 4.5292 0.0333
ivhx3 1 -0.7046 0.2620 7.2326 0.0072
RACE 1 0.6258 0.2672 5.4865 0.0192
TREAT 1 0.4669 0.2050 5.1889 0.0227
SITE 1 0.5310 0.2550 4.3344 0.0373
agendrgfp1 1 -0.0140 0.00607 5.3171 0.0211
racesite 1 -1.3700 0.5312 6.6530 0.0099
[output omitted]
Hosmer and Lemeshow Goodness-of-Fit Test
Chi-Square DF Pr > ChiSq
7.3579 8 0.4986
*Column 5 of Table 5.9;
proc logistic data=uis54 desc;
where covpat not in (468);
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site
agendrgfp1 racesite / aggregate lackfit scale = 1;
run;
[output omitted]
Deviance and Pearson Goodness-of-Fit Statistics
Criterion Value DF Value/DF Pr > ChiSq
Deviance 526.9371 509 1.0352 0.2821
Pearson 511.5712 509 1.0051 0.4596
Number of unique profiles: 520
[output omitted]
Analysis of Maximum Likelihood Estimates
Standard Wald
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -7.0471 1.2379 32.4064 <.0001
AGE 1 0.1223 0.0293 17.4026 <.0001
ndrgfp1 1 1.7645 0.4173 17.8804 <.0001
ndrgfp2 1 0.4501 0.1182 14.4946 0.0001
ivhx2 1 -0.6393 0.2990 4.5700 0.0325
ivhx3 1 -0.7455 0.2636 7.9959 0.0047
RACE 1 0.6437 0.2660 5.8549 0.0155
TREAT 1 0.4507 0.2045 4.8552 0.0276
SITE 1 0.5164 0.2553 4.0920 0.0431
agendrgfp1 1 -0.0173 0.00627 7.5916 0.0059
racesite 1 -1.3784 0.5318 6.7182 0.0095
[output omitted]
Hosmer and Lemeshow Goodness-of-Fit Test
Chi-Square DF Pr > ChiSq
9.0942 8 0.3344
*Column 6 of Table 5.9;
proc logistic data=uis54 desc;
where covpat not in (31, 477, 105, 468);
model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site
agendrgfp1 racesite / aggregate lackfit scale=1;
run;
[output omitted]
Deviance and Pearson Goodness-of-Fit Statistics
Criterion Value DF Value/DF Pr > ChiSq
Deviance 511.1110 506 1.0101 0.4282
Pearson 482.6328 506 0.9538 0.7658
Number of unique profiles: 517
[output omitted]
Analysis of Maximum Likelihood Estimates
Standard Wald
Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -7.7998 1.2995 36.0240 <.0001
AGE 1 0.1376 0.0306 20.2463 <.0001
ndrgfp1 1 2.0425 0.4430 21.2590 <.0001
ndrgfp2 1 0.5253 0.1228 18.2911 <.0001
ivhx2 1 -0.7017 0.3043 5.3167 0.0211
ivhx3 1 -0.7962 0.2678 8.8386 0.0029
RACE 1 0.5454 0.2730 3.9904 0.0458
TREAT 1 0.5253 0.2084 6.3525 0.0117
SITE 1 0.5042 0.2584 3.8069 0.0510
agendrgfp1 1 -0.0204 0.00677 9.0996 0.0026
racesite 1 -1.2509 0.5387 5.3931 0.0202
[output omitted]
Hosmer and Lemeshow Goodness-of-Fit Test
Chi-Square DF Pr > ChiSq
6.8554 8 0.5523
page 189 Table 5.10 Estimated coefficients, standard errors, z-scores, two-tailed p-values and 95% confidence intervals for the final logistic regression model for the UIS (n=575).
proc logistic data=uis51 desc; model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite / clparm=both; run;The LOGISTIC Procedure
Model Information
Data Set WORK.UIS51 Response Variable DFREE Number of Response Levels 2 Number of Observations 575 Link Function Logit Optimization Technique Fisher's scoring
Response Profile
Ordered Total Value DFREE Frequency
1 1 147 2 0 428
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 655.729 619.963 SC 660.083 667.861 -2 Log L 653.729 597.963
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 55.7660 10 <.0001 Score 52.0723 10 <.0001 Wald 47.2784 10 <.0001
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001 AGE 1 0.1166 0.0289 16.3137 <.0001 ndrgfp1 1 1.6687 0.4071 16.8000 <.0001 ndrgfp2 1 0.4336 0.1169 13.7585 0.0002 IVHX2 1 -0.6346 0.2987 4.5134 0.0336 IVHX3 1 -0.7049 0.2616 7.2623 0.0070 RACE 1 0.6841 0.2641 6.7074 0.0096 TREAT 1 0.4349 0.2038 4.5559 0.0328 SITE 1 0.5162 0.2549 4.1013 0.0429 agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113 racesite 1 -1.4294 0.5298 7.2799 0.0070
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
AGE 1.124 1.062 1.189 ndrgfp1 5.306 2.389 11.784 ndrgfp2 1.543 1.227 1.940 IVHX2 0.530 0.295 0.952 IVHX3 0.494 0.296 0.825 RACE 1.982 1.181 3.326 TREAT 1.545 1.036 2.303 SITE 1.676 1.017 2.761 agendrgfp1 0.985 0.973 0.997 racesite 0.239 0.085 0.676
Association of Predicted Probabilities and Observed Responses
Percent Concordant 69.7 Somers' D 0.398 Percent Discordant 29.9 Gamma 0.399 Percent Tied 0.4 Tau-a 0.152 Pairs 62916 c 0.699
Profile Likelihood Confidence Interval for Parameters
Parameter Estimate 95% Confidence Limits
Intercept -6.8429 -9.3201 -4.5308 AGE 0.1166 0.0611 0.1746 ndrgfp1 1.6687 0.8956 2.4954 ndrgfp2 0.4336 0.2088 0.6678
The LOGISTIC Procedure
Profile Likelihood Confidence Interval for Parameters
Parameter Estimate 95% Confidence Limits
IVHX2 -0.6346 -1.2332 -0.0590 IVHX3 -0.7049 -1.2234 -0.1960 RACE 0.6841 0.1638 1.2013 TREAT 0.4349 0.0373 0.8372 SITE 0.5162 0.0143 1.0153 agendrgfp1 -0.0153 -0.0276 -0.00382 racesite -1.4294 -2.5080 -0.4174
Wald Confidence Interval for Parameters
Parameter Estimate 95% Confidence Limits
Intercept -6.8429 -9.2326 -4.4532 AGE 0.1166 0.0600 0.1732 ndrgfp1 1.6687 0.8708 2.4667 ndrgfp2 0.4336 0.2045 0.6627 IVHX2 -0.6346 -1.2201 -0.0491 IVHX3 -0.7049 -1.2176 -0.1922 RACE 0.6841 0.1664 1.2018 TREAT 0.4349 0.0356 0.8343 SITE 0.5162 0.0166 1.0157 agendrgfp1 -0.0153 -0.0271 -0.00346 racesite -1.4294 -2.4677 -0.3911
page 190 Table 5.11 Estimated odds ratios and 95% confidence intervals for treatment and history of IV drug use in the UIS (N = 575).
proc logistic data=uis51 desc; model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite; run;The LOGISTIC Procedure
Model Information
Data Set WORK.UIS51 Response Variable DFREE Number of Response Levels 2 Number of Observations 575 Link Function Logit Optimization Technique Fisher's scoring
Response Profile
Ordered Total Value DFREE Frequency
1 1 147 2 0 428
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 655.729 619.963 SC 660.083 667.861 -2 Log L 653.729 597.963
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 55.7660 10 <.0001 Score 52.0723 10 <.0001 Wald 47.2784 10 <.0001
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -6.8429 1.2193 31.4989 <.0001 AGE 1 0.1166 0.0289 16.3137 <.0001 ndrgfp1 1 1.6687 0.4071 16.8000 <.0001 ndrgfp2 1 0.4336 0.1169 13.7585 0.0002 IVHX2 1 -0.6346 0.2987 4.5134 0.0336 IVHX3 1 -0.7049 0.2616 7.2623 0.0070 RACE 1 0.6841 0.2641 6.7074 0.0096 TREAT 1 0.4349 0.2038 4.5559 0.0328 SITE 1 0.5162 0.2549 4.1013 0.0429 agendrgfp1 1 -0.0153 0.00603 6.4177 0.0113 racesite 1 -1.4294 0.5298 7.2799 0.0070
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
AGE 1.124 1.062 1.189 ndrgfp1 5.306 2.389 11.784 ndrgfp2 1.543 1.227 1.940 IVHX2 0.530 0.295 0.952 IVHX3 0.494 0.296 0.825 RACE 1.982 1.181 3.326 TREAT 1.545 1.036 2.303 SITE 1.676 1.017 2.761 agendrgfp1 0.985 0.973 0.997 racesite 0.239 0.085 0.676
Association of Predicted Probabilities and Observed Responses
Percent Concordant 69.7 Somers' D 0.398 Percent Discordant 29.9 Gamma 0.399 Percent Tied 0.4 Tau-a 0.152 Pairs 62916 c 0.699
page 192 Table 5.12 Estimated odds ratios and 95% confidence intervals for race within site in the UIS (n = 575).
proc genmod data=uis51 descending; model dfree = age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite / dist=bin link=logit waldci; estimate 'race = other, site = A' race 1 /exp; estimate 'race = other, site = B' race 1 racesite 1 /exp; run;The GENMOD Procedure
Model Information
Data Set WORK.UIS51 Distribution Binomial Link Function Logit Dependent Variable DFREE Observations Used 575 Probability Modeled Pr( DFREE = 1 )
Response Profile
Ordered Ordered Level Value Count
1 0 428 2 1 147
Parameter Information
Parameter Effect
Prm1 Intercept Prm2 AGE Prm3 ndrgfp1 Prm4 ndrgfp2 Prm5 IVHX2 Prm6 IVHX3 Prm7 RACE Prm8 TREAT Prm9 SITE Prm10 agendrgfp1 Prm11 racesite
Criteria For Assessing Goodness Of Fit
Criterion DF Value Value/DF
Deviance 564 597.9629 1.0602 Scaled Deviance 564 597.9629 1.0602 Pearson Chi-Square 564 580.7351 1.0297 Scaled Pearson X2 564 580.7351 1.0297 Log Likelihood -298.9815
Algorithm converged. The GENMOD Procedure
Analysis Of Parameter Estimates
Standard Wald 95% Confidence Chi- Parameter DF Estimate Error Limits Square Pr > ChiSq
Intercept 1 -6.8439 1.2193 -9.2337 -4.4540 31.50 <.0001 AGE 1 0.1166 0.0289 0.0600 0.1732 16.32 <.0001 ndrgfp1 1 1.6690 0.4072 0.8710 2.4670 16.80 <.0001 ndrgfp2 1 0.4337 0.1169 0.2046 0.6628 13.76 0.0002 IVHX2 1 -0.6346 0.2987 -1.2201 -0.0492 4.51 0.0336 IVHX3 1 -0.7049 0.2616 -1.2176 -0.1923 7.26 0.0070 RACE 1 0.6841 0.2641 0.1664 1.2018 6.71 0.0096 TREAT 1 0.4349 0.2038 0.0356 0.8343 4.56 0.0328 SITE 1 0.5162 0.2549 0.0166 1.0158 4.10 0.0428 agendrgfp1 1 -0.0153 0.0060 -0.0271 -0.0035 6.42 0.0113 racesite 1 -1.4295 0.5298 -2.4678 -0.3911 7.28 0.0070 Scale 0 1.0000 0.0000 1.0000 1.0000
NOTE: The scale parameter was held fixed.
Contrast Estimate Results
Standard Chi- Label Estimate Error Alpha Confidence Limits Square Pr > ChiSq
race = other, site = A 0.6841 0.2641 0.05 0.1664 1.2018 6.71 0.0096 Exp(race = other, site = A) 1.9820 0.5235 0.05 1.1811 3.3261 race = other, site = B -0.7454 0.4636 0.05 -1.6540 0.1633 2.58 0.1079 Exp(race = other, site = B) 0.4746 0.2200 0.05 0.1913 1.1774
page 194 Figure 5.9 Estimated odds ratio and 95% confidence limits for a five-year increase in age based on the model in Table 5.10.
NOTE: We were unable to reproduce this graph.
page 197 Figure 5.10 Estimated odds ratios and 95% confidence limits for an increase of one drug treatment from the plotted value of NDRGTX for a subject of age (a) 20, (b) 25, (c) 30 and (d) 35.
NOTE: We were unable to reproduce this graph.
page 199 Figure 5.11 Estimated odds ratios and 95% confidence limits comparing zero, two, three up to 10 previous drug treatments to one previous treatment for a subject of age (a) 20, (b) 25, (c) 30 and (d) 35.
NOTE: We were unable to reproduce this graph.