options nocenter nodate;
Inputting the Programming Task data, table 14.1, p. 576.
data ch14tab01;
input x y ;
label x = 'Experience'
y = 'Success';
cards;
14 0 0.310262
29 0 0.835263
6 0 0.109996
25 1 0.726602
18 1 0.461837
4 0 0.082130
18 0 0.461837
12 0 0.245666
22 1 0.620812
6 0 0.109996
30 1 0.856299
11 0 0.216980
30 1 0.856299
5 0 0.095154
20 1 0.542404
13 0 0.276802
9 0 0.167100
32 1 0.891664
24 0 0.693379
13 1 0.276802
19 0 0.502134
4 0 0.082130
28 1 0.811825
22 1 0.620812
8 1 0.145815
;
run;
Logistic Regression, table 14.1, p. 576.
proc logistic data = ch14tab01 descending; model y = x; output out = temp resdev=devresidual p = fittedp; run; proc print data = temp; var x y fittedp devresidual; run;
The LOGISTIC ProcedureModel Information
Data Set WORK.CH14TAB01 Response Variable y Success Number of Response Levels 2 Number of Observations 25 Link Function Logit Optimization Technique Fisher’s scoring
Response Profile
Ordered Total Value y Frequency
1 1 11 2 0 14
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 36.296 29.425 SC 37.515 31.862 -2 Log L 34.296 25.425
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 8.8719 1 0.0029 Score 7.9742 1 0.0047 Wald 6.1760 1 0.0129
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -3.0597 1.2594 5.9029 0.0151 x 1 0.1615 0.0650 6.1760 0.0129
The LOGISTIC Procedure
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
x 1.175 1.035 1.335
Association of Predicted Probabilities and Observed Responses
Percent Concordant 82.5 Somers’ D 0.662 Percent Discordant 16.2 Gamma 0.671 Percent Tied 1.3 Tau-a 0.340 Pairs 154 c 0.831 Obs x y fittedp devresidual
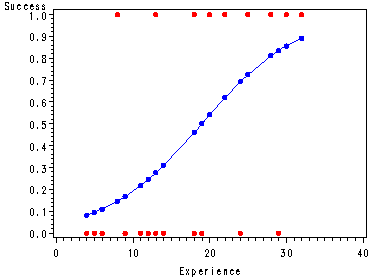
1 14 0 0.31026 -0.86191 2 29 0 0.83526 -1.89916 3 6 0 0.11000 -0.48276 4 25 1 0.72660 0.79922 5 18 1 0.46184 1.24302 6 4 0 0.08213 -0.41400 7 18 0 0.46184 -1.11319 8 12 0 0.24567 -0.75089 9 22 1 0.62081 0.97645 10 6 0 0.11000 -0.48276 11 30 1 0.85630 0.55702 12 11 0 0.21698 -0.69942 13 30 1 0.85630 0.55702 14 5 0 0.09515 -0.44719 15 20 1 0.54240 1.10611 16 13 0 0.27680 -0.80507 17 9 0 0.16710 -0.60472 18 32 1 0.89166 0.47889 19 24 0 0.69338 -1.53762 20 13 1 0.27680 1.60278 21 19 0 0.50213 -1.18104 22 4 0 0.08213 -0.41400 23 28 1 0.81182 0.64571 24 22 1 0.62081 0.97645 25 8 1 0.14582 1.96235
Fig. 14.3, p. 576.
proc sort data = temp; by x; run; goptions reset = all; symbol1 c=red v=dot h = .8 ; symbol2 c=blue v=dot h=.8 i=join; proc gplot data = temp; plot y*x fittedp*x / overlay; run; quit; goptions reset = all;
Inputting Coupon Effectiveness data, Table 14.2, p. 579.
data ch14tab02;
input x n r p;
label x = 'Reduction'
n = 'no. households'
r = 'coupons redeemed'
p = 'proportion of coupons redeemed';
cards;
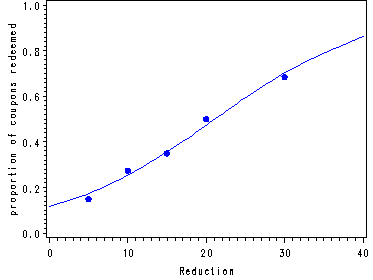
5 200 30 .150
10 200 55 .275
15 200 70 .350
20 200 100 .500
30 200 137 .685
;
run;
Fig. 14.4, p. 579.
In order to implement logistic regression using proportions it is necessary to use proc genmod and specify the distribution and the link function. The parameter estimates in the output correspond to the fitted response function (14.28) at the bottom of p. 578.
proc genmod data=ch14tab02; model r/n = x / dist = bin link = logit lrci; output out=temp p=predicted; run;
The GENMOD ProcedureModel Information
Data Set WORK.CH14TAB02 Distribution Binomial Link Function Logit Response Variable (Events) r coupons redeemed Response Variable (Trials) n no. households Observations Used 5 Number Of Events 392 Number Of Trials 1000
Criteria For Assessing Goodness Of Fit
Criterion DF Value Value/DF
Deviance 3 2.1668 0.7223 Scaled Deviance 3 2.1668 0.7223 Pearson Chi-Square 3 2.1486 0.7162 Scaled Pearson X2 3 2.1486 0.7162 Log Likelihood -595.9863
Algorithm converged.
Analysis Of Parameter Estimates
Standard Likelihood Ratio 95% Chi- Parameter DF Estimate Error Confidence Limits Square Pr > ChiSq
Intercept 1 -2.0443 0.1610 -2.3655 -1.7340 161.28 <.0001 x 1 0.0968 0.0085 0.0803 0.1139 128.29 <.0001 Scale 0 1.0000 0.0000 1.0000 1.0000
NOTE: The scale parameter was held fixed.
Fig. 14.4, p. 579. Fitted values for X=0 and X=40 have been added in order for the fitted curve to extend beyond the range of the X variable in the data set.
data extra; if _n_ = 1 then do; predicted = exp(-2.04435) / (1+ exp(-2.04435) ); x=0; output; predicted = exp(-2.04435 + 0.096834*40) / (1+ exp(-2.04435 + 0.096834*40) ); x=40; output; end; set temp; output; run; proc sort data = extra; by x; run; symbol1 v=dot c=blue; symbol2 i=spline v=none c=blue; axis1 label=(angle = 90 h = 1) order=(0 to 1.0 by .2); axis2 order=(0 to 40 by 10); proc gplot data = extra; plot (p predicted)*x / overlay vaxis=axis1 haxis=axis2; run; quit; goptions reset = all;
Inputting the Disease Outbreak data, table 14.3, p. 583.
data ch14tab03;
input id x1 socio x4 y x5;
label id = 'case'
x1 = 'age'
socio = 'socioeconomic status'
x4 = 'sector'
y = 'Disease status'
x5 = 'savings';
cards;
1 33 1 1 0 1
2 35 1 1 0 1
3 6 1 1 0 0
4 60 1 1 0 1
5 18 3 1 1 0
6 26 3 1 0 0
7 6 3 1 0 0
8 31 2 1 1 1
9 26 2 1 1 0
10 37 2 1 0 0
11 23 1 1 0 0
12 23 1 1 0 0
13 27 1 1 0 1
14 9 1 1 1 1
15 37 1 2 1 1
16 22 1 2 1 1
17 67 1 2 1 1
18 8 1 2 0 1
19 6 1 2 1 1
20 15 1 2 1 1
21 21 2 2 1 1
22 32 2 2 1 1
23 16 1 2 1 1
24 11 2 2 0 0
25 14 3 2 0 0
26 9 2 2 0 0
27 18 2 2 0 0
28 2 3 1 0 0
29 61 3 1 0 1
30 20 3 1 0 0
31 16 3 1 0 0
32 9 2 1 0 0
33 35 2 1 0 1
34 4 1 1 0 1
35 44 3 2 0 0
36 11 3 2 1 0
37 3 2 2 0 1
38 6 3 2 0 0
39 17 2 2 1 0
40 1 3 2 0 1
41 53 2 2 1 1
42 13 1 2 1 0
43 24 1 2 0 0
44 70 1 2 1 1
45 16 3 2 1 1
46 12 2 2 0 1
47 20 3 2 1 1
48 65 3 2 0 1
49 40 2 2 1 0
50 38 2 2 1 1
51 68 2 2 1 1
52 74 1 2 1 1
53 14 1 2 1 1
54 27 1 2 1 1
55 31 1 2 0 1
56 18 1 2 0 1
57 39 1 2 0 0
58 50 1 2 0 1
59 31 1 2 0 1
60 61 1 2 0 1
61 18 3 1 0 0
62 5 3 1 0 0
63 2 3 1 0 1
64 16 3 1 0 0
65 59 3 1 1 1
66 22 3 1 0 0
67 24 1 1 0 1
68 30 1 1 0 1
69 46 1 1 0 1
70 28 1 1 0 0
71 27 1 1 0 1
72 27 1 1 1 0
73 28 1 1 0 1
74 52 1 1 1 1
75 11 3 1 0 1
76 6 2 1 0 1
77 46 3 1 0 0
78 20 2 1 1 1
79 3 1 1 0 1
80 18 2 1 0 0
81 25 2 1 0 0
82 6 3 1 0 1
83 65 3 1 1 1
84 51 3 1 0 1
85 39 2 1 0 1
86 8 1 1 0 1
87 8 2 1 0 0
88 14 3 1 0 0
89 6 3 1 0 0
90 6 3 1 0 1
91 7 3 1 0 0
92 4 3 1 0 0
93 8 3 1 0 0
94 9 2 1 0 0
95 32 3 1 1 0
96 19 3 1 0 0
97 11 3 1 0 0
98 35 3 1 0 0
;
run;
Creating the dummy variables for socioeconomic status.
data ch14tb03a; set ch14tab03; x2 = 0; if socio = 2 then x2 = 1; x3 = 0; if socio = 3 then x3 = 1; run;
Table 14.4, p. 584. It is the option covb in the model statement that gives us part b of the table.
Note: The estimate for the intercept is different from the book perhaps because the authors used a slightly different algorithm. However, it is usually the odds ratio of the other parameters estimates that are of interest and they are the same as in the book.
proc logistic data = ch14tb03a descending; model y = x1 x2 x3 x4/ covb; run;
The LOGISTIC ProcedureModel Information
Data Set WORK.CH14TB03A Response Variable y Disease status Number of Response Levels 2 Number of Observations 98 Link Function Logit Optimization Technique Fisher’s scoring
Response Profile
Ordered Total Value y Frequency 1 1 31 2 0 67
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 124.318 111.054 SC 126.903 123.979 -2 Log L 122.318 101.054
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 21.2635 4 0.0003 Score 20.4067 4 0.0004 Wald 16.6437 4 0.0023
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -3.8874 0.9955 15.2496 <.0001 x1 1 0.0297 0.0135 4.8535 0.0276 x2 1 0.4088 0.5990 0.4657 0.4950 x3 1 -0.3051 0.6041 0.2551 0.6135 x4 1 1.5746 0.5016 9.8543 0.0017
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
x1 1.030 1.003 1.058 x2 1.505 0.465 4.868 x3 0.737 0.226 2.408 x4 4.829 1.807 12.907
Association of Predicted Probabilities and Observed Responses
Percent Concordant 77.5 Somers’ D 0.554 Percent Discordant 22.1 Gamma 0.556 Percent Tied 0.3 Tau-a 0.242 Pairs 2077 c 0.777
Estimated Covariance Matrix
Variable Intercept x1 x2 x3 x4
Intercept 0.990945 -0.00605 -0.19645 -0.26324 -0.41483 x1 -0.00605 0.000182 0.00115 0.000732 0.000338 x2 -0.19645 0.00115 0.358793 0.148217 0.012887 x3 -0.26324 0.000732 0.148217 0.364944 0.062267 x4 -0.41483 0.000338 0.012887 0.062267 0.251609
Testing multiple parameters, p. 589.
In SAS testing linear hypotheses about the regression coefficients is done using a Wald test. To use the built in SAS option just add test statements for all the hypothesis that needs to be tested. The partial deviance can be used by running the full and reduced model for each hypothesis and then taking each model and comparing this difference to the appropriate chi-square distribution.
proc logistic data = ch14tb03a descending; model y = x1 x2 x3 x4; test: test x1=0; run; proc logistic data = ch14tb03a descending; model y = x2 x3 x4; run;
The LOGISTIC ProcedureModel Information
Data Set WORK.CH14TB03A Response Variable y Disease status Number of Response Levels 2 Number of Observations 98 Link Function Logit Optimization Technique Fisher’s scoring
Response Profile
Ordered Total Value y Frequency
1 1 31 2 0 67
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 124.318 111.054 SC 126.903 123.979 -2 Log L 122.318 101.054
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 21.2635 4 0.0003 Score 20.4067 4 0.0004 Wald 16.6437 4 0.0023
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -3.8874 0.9955 15.2496 <.0001 x1 1 0.0297 0.0135 4.8535 0.0276 x2 1 0.4088 0.5990 0.4657 0.4950 x3 1 -0.3051 0.6041 0.2551 0.6135 x4 1 1.5746 0.5016 9.8543 0.0017
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
x1 1.030 1.003 1.058 x2 1.505 0.465 4.868 x3 0.737 0.226 2.408 x4 4.829 1.807 12.907
Association of Predicted Probabilities and Observed Responses
Percent Concordant 77.5 Somers’ D 0.554 Percent Discordant 22.1 Gamma 0.556 Percent Tied 0.3 Tau-a 0.242 Pairs 2077 c 0.777
Linear Hypotheses Testing Results
Wald Label Chi-Square DF Pr > ChiSq
test 4.8535 1 0.0276
The LOGISTIC Procedure
Model Information
Data Set WORK.CH14TB03A Response Variable y Disease status Number of Response Levels 2 Number of Observations 98 Link Function Logit Optimization Technique Fisher’s scoring
Response Profile
Ordered Total Value y Frequency
1 1 31 2 0 67
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 124.318 114.204 SC 126.903 124.544 -2 Log L 122.318 106.204
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 16.1139 3 0.0011 Score 15.8641 3 0.0012 Wald 14.2743 3 0.0026
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -3.0595 0.8639 12.5427 0.0004 x2 1 0.2351 0.5752 0.1670 0.6828 x3 1 -0.4779 0.5829 0.6721 0.4123 x4 1 1.6203 0.4857 11.1289 0.0008
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
x2 1.265 0.410 3.906 x3 0.620 0.198 1.944 x4 5.055 1.951 13.095
Association of Predicted Probabilities and Observed Responses
Percent Concordant 65.8 Somers’ D 0.465 Percent Discordant 19.3 Gamma 0.546 Percent Tied 14.9 Tau-a 0.203 Pairs 2077 c 0.733
Creating all the interactions to be tested.
data ch14tb03b; set ch14tb03a; x1x2 = x1*x2; x1x3 = x1*x3; x1x4 = x1*x4; x2x4 = x2*x4; x3x4 = x3*x4; run;
Testing the interactions, p. 589.
proc logistic data = ch14tb03b descending; model y = x1-x4 x1x2 x1x3 x1x4 x2x4 x3x4; test: test x1x2=x1x3= x1x4= x2x4= x3x4=0; run;
The LOGISTIC ProcedureModel Information
Data Set WORK.CH14TB03B Response Variable y Disease status Number of Response Levels 2 Number of Observations 98 Link Function Logit Optimization Technique Fisher’s scoring
Response Profile
Ordered Total Value y Frequency
1 1 31 2 0 67
Model Convergence Status Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 124.318 113.996 SC 126.903 139.846 -2 Log L 122.318 93.996
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 28.3217 9 0.0008 Score 25.6302 9 0.0023 Wald 17.9067 9 0.0363
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -5.5161 2.2471 6.0260 0.0141 x1 1 0.0646 0.0583 1.2294 0.2675 x2 1 -1.7862 3.0841 0.3354 0.5625 x3 1 0.2955 2.2550 0.0172 0.8957 x4 1 2.9796 1.2481 5.6988 0.0170 x1x2 1 0.1054 0.0559 3.5514 0.0595 x1x3 1 0.0140 0.0316 0.1952 0.6586 x1x4 1 -0.0342 0.0309 1.2231 0.2688 x2x4 1 -0.3094 1.4409 0.0461 0.8300 x3x4 1 -0.7396 1.2489 0.3507 0.5537
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
x1 1.067 0.952 1.196 x2 0.168 <0.001 70.702 x3 1.344 0.016 111.632 x4 19.680 1.705 227.221 x1x2 1.111 0.996 1.240 x1x3 1.014 0.953 1.079 x1x4 0.966 0.910 1.027 x2x4 0.734 0.044 12.363 x3x4 0.477 0.041 5.519
Association of Predicted Probabilities and Observed Responses
Percent Concordant 80.4 Somers’ D 0.610 Percent Discordant 19.4 Gamma 0.612 Percent Tied 0.3 Tau-a 0.267 Pairs 2077 c 0.805
Linear Hypotheses Testing Results
Wald Label Chi-Square DF Pr > ChiSq
test 5.9413 5 0.3120
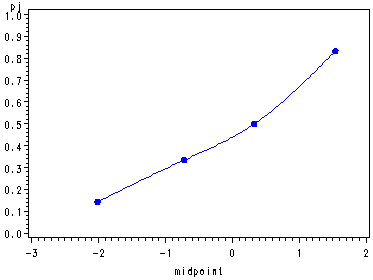
Example 1, p. 591 and Fig. 14.5, p. 592.
Invoking the macro diag_plot.
Note: The macro splits the observations into groups with an equal number of observations except for the last group therefore they may not match the groups in the book since they are not the same size as those in the book. Also, the results from the logistic regression produced by the macro has been omitted.
%include "c:neter/sas/examples/alsm/diag_plot.sas"; %diag_plot(ch14tab01, y, x, 4);
Obs class min max midpoint n pj1 1 -2.41375 -1.60632 -2.01004 7 0.14286 2 2 -1.28335 -0.15295 -0.71815 6 0.33333 3 3 -0.15295 0.81597 0.33151 6 0.50000 4 4 0.97745 2.10785 1.54265 6 0.83333
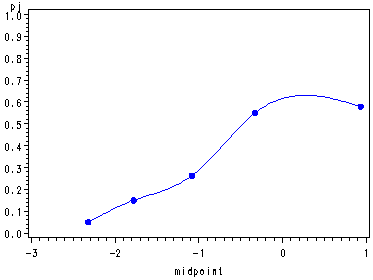
Example 2, p. 591 and Fig. 14.6, p. 592.
Invoking the macro diag_plot again and again the results from the logistic regression produced by the macro has been omitted.
%diag_plot(ch14tb03a, y, x1 x2 x3 x4, 5);
Obs class min max midpoint n pj1 1 -2.55835 -2.08241 -2.32038 20 0.05000 2 2 -2.07476 -1.47983 -1.77729 20 0.15000 3 3 -1.42033 -0.74386 -1.08210 19 0.26316 4 4 -0.71601 0.06505 -0.32548 20 0.55000 5 5 0.17633 1.69341 0.93487 19 0.57895
Table 14.5, p. 594.
Note: The numbers are not exactly the same as those in the book most probably due to rounding errors. Only the output from the final print procedure has been included in the results.
proc logistic data = ch14tb03a descending;
model y = x1 x2 x3 x4 ;
output out=temp p = pi;
run;
data temp;
set temp;
pihat = log( pi / (1 - pi) );
run;
proc sort data = temp;
by pihat;
run;
data temp;
set temp nobs=total;
class = .;
class = int( ( _n_ - 1 )/( total/5 ) ) +1;
run;
proc sql;
create table temp1 as
select *, max(pihat) as max, min(pihat) as min, sum(pi) as Ej1, count(pi) as n,
sum(y) as Oj1, count(pi) - sum(pi) as Ej0, count(pi) - sum(y) as Oj0
from temp
group by class;
quit;
proc sort data = temp1 (keep = class n min max Oj0 Ej0 Oj1 Ej1);
by class ;
run;
data temp1;
set temp1;
by class;
if first.class;
run;
proc print data=temp1;
var class min max n Oj0 Ej0 Oj1 Ej1;
run;
Obs class min max n Oj0 Ej0 Oj1 Ej11 1 -2.55835 -2.08241 20 19 18.1952 1 1.8048 2 2 -2.07476 -1.47983 20 17 16.9072 3 3.0928 3 3 -1.42033 -0.74386 19 14 14.0400 5 4.9600 4 4 -0.71601 0.06505 20 9 11.5587 11 8.4413 5 5 0.17633 1.69341 19 8 6.2976 11 12.7024
Index plots, including the RESDEV (Residual deviance) plot which is the same as Fig. 14.7, p. 596.
proc logistic data = ch14tab01 desc; model y = x / iplots; run;
The LOGISTIC ProcedureModel Information
Data Set WORK.CH14TAB01 Response Variable y Success Number of Response Levels 2 Number of Observations 25 Link Function Logit Optimization Technique Fisher’s scoring
Response Profile
Ordered Total Value y Frequency
1 1 11 2 0 14
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 36.296 29.425 SC 37.515 31.862 -2 Log L 34.296 25.425
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 8.8719 1 0.0029 Score 7.9742 1 0.0047 Wald 6.1760 1 0.0129
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -3.0597 1.2594 5.9029 0.0151 x 1 0.1615 0.0650 6.1760 0.0129
The LOGISTIC Procedure
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
x 1.175 1.035 1.335
Association of Predicted Probabilities and Observed Responses
Percent Concordant 82.5 Somers’ D 0.662 Percent Discordant 16.2 Gamma 0.671 Percent Tied 1.3 Tau-a 0.340 Pairs 154 c 0.831
The LOGISTIC Procedure
—–+————–+————–+————–+————–+————–+—— RESCHI | | P 4 + + e | | a | | r | * | s 2 + + o | * | n | * * * * | | * * * * * | R 0 + + e | * * * * * * * * * * | s | * * | i | * | d -2 + + u | * | a | | l | | -4 + + | | —–+————–+————–+————–+————–+————–+—— 0 5 10 15 20 25
Case Number INDEX
—–+————–+————–+————–+————–+————–+—— 2 + * + D | | e | * | v | * | i RESDEV | * | a | * * * | n | * * * | c | * | e | | 0 + + R | | e | * * * * * | s | * * * | i | * * | d | * * | u | | a | * | l | | -2 + * + —–+————–+————–+————–+————–+————–+—— 0 5 10 15 20 25
Case Number INDEX
The LOGISTIC Procedure
——+————–+————–+————–+————–+————–+——- 0.12 + + | | | | H | | a | * * * | t | * * | 0.10 + + D | | i | | a H | * | g | | o | * | n 0.08 + * * * * * * * + a | | l | * * * | | * | | * * | | * * | 0.06 + * * * + ——+————–+————–+————–+————–+————–+——- 0 5 10 15 20 25
Case Number INDEX
—–+————–+————–+————–+————–+————–+—– 1.0 + + I | | n | | t | * | e | | r | | c 0.5 + + e | * | p | * | t DFBETA0 | | | * | D | * | f 0.0 + * * * + B | * * * * * * * * * * * * | e | * * * * * | t | | a | | | | -0.5 + + —–+————–+————–+————–+————–+————–+—– 0 5 10 15 20 25
Case Number INDEX
The LOGISTIC Procedure
—–+————–+————–+————–+————–+————–+—– 0.5 + + | | | | | | | * * * * | x | * * * * * * * * * * * * * * | 0.0 + * * * + D | | f | | B DFBETA1 | * | e | * | t | | a -0.5 + + | * | | * | | | | | | | -1.0 + + —–+————–+————–+————–+————–+————–+—– 0 5 10 15 20 25
Case Number INDEX
——+————–+————–+————–+————–+————–+——- C 0.75 + + o | | n | * | f | | i | | d | * | e 0.50 + + n | | c | | e C | | | | I | | n 0.25 + + t | * * | e | | r | | v | * * | a | * * * * * * * * * * * * | l 0.00 + * * * * * * * + ——+————–+————–+————–+————–+————–+——- D 0 5 10 15 20 25 i Case Number INDEX
The LOGISTIC Procedure
——+————–+————–+————–+————–+————–+——- C 0.6 + * + o | | n | | f | * | i | | d | | e 0.4 + + n | | c | | e CBAR | | | | I | | n 0.2 + * * + t | | e | | r | | v | * * * * | a | * * * * * * * * * * * | l 0.0 + * * * * * * + ——+————–+————–+————–+————–+————–+——- D 0 5 10 15 20 25 i Case Number INDEX
—–+————–+————–+————–+————–+————–+—— 6 + + | | D | | e | | l | | t | * * | a 4 + + | | D | | e DIFDEV | | v | * * | i | | a 2 + + n | * | c | * * * | e | * * | | * * * * * | | * * * * * * * * * * | 0 + + —–+————–+————–+————–+————–+————–+—— 0 5 10 15 20 25
Case Number INDEX
The LOGISTIC Procedure
—-+————–+————–+————–+————–+————–+—– DIFCHISQ | | 8 + + D | | e | | l | * | t 6 + + a | * | | | C | | h 4 + + i | | S | * | q | * | u 2 + + a | | r | * * * * | e | * * * * * * * * | 0 + * * * * * * * * * + | | —-+————–+————–+————–+————–+————–+—– 0 5 10 15 20 25
Case Number INDEX
Predicting mean responses with confidence interval, example p. 604-605.
The output contains the point estimate of the logit mean response as phat, the confidence limits for the logit mean response as lower1 and upper1, the point estimate for the mean response as p, and finally, the confidence interval for the mean response as lower and upper. The output from the proc logistic is not shown.
data ch14tb03b;
if _n_ = 1 then do;
id = 99; x1=10; x2=0; x3=1; x4=1;
end;
output;
set ch14tb03a;
run;
proc logistic data = ch14tb03b desc;
model y = x1 x2 x3 x4;
output out=temp p=p upper=upper lower=lower;
run;
data temp;
set temp;
lower1 = log(lower/ (1-lower) ) ;
upper1 = log(upper / (1-upper) );
phat = log(p / (1-p) );
run;
proc print data = temp;
where id = 99;
var phat lower1 upper1 p lower upper;
run;
Obs phat lower1 upper1 p lower upper1 -2.32038 -3.38397 -1.25679 0.089449 0.032800 0.22153
Table 14.7, p. 607.
The table produced by SAS is very different from the table in the book. The book uses the list of predicted fitted values and then compares them to a specified cutoff point. SAS does not use this method because when you classify binary data and the observations that are used to fit the model are also used to estimate the classification error then the resulting error-count estimate is biased. One way to reduce the bias is to remove the observation to be classified and re-estimate the parameters of the model and then classify the observation based on the parameter estimates based on the smaller dataset (without the observation to be classified). In order to increase efficiency SAS uses a one-step approximation of the parameter estimates based on the smaller dataset (without the observation to be classified). For the details of the one-step approximation please refer to the manual under Proc Logistic Classification Table.
proc logistic data = ch14tb03a desc; model y = x1 x2 x3 x4/ ctable; output out=temp p=p; run;
The LOGISTIC ProcedureModel Information
Data Set WORK.CH14TB03A Response Variable y Disease status Number of Response Levels 2 Number of Observations 98 Link Function Logit Optimization Technique Fisher’s scoring
Response Profile
Ordered Total Value y Frequency
1 1 31 2 0 67
Model Convergence Status
Convergence criterion (GCONV=1E-8) satisfied.
Model Fit Statistics
Intercept Intercept and Criterion Only Covariates
AIC 124.318 111.054 SC 126.903 123.979 -2 Log L 122.318 101.054
Testing Global Null Hypothesis: BETA=0
Test Chi-Square DF Pr > ChiSq
Likelihood Ratio 21.2635 4 0.0003 Score 20.4067 4 0.0004 Wald 16.6437 4 0.0023
The LOGISTIC Procedure
Analysis of Maximum Likelihood Estimates
Standard Parameter DF Estimate Error Chi-Square Pr > ChiSq
Intercept 1 -3.8874 0.9955 15.2496 <.0001 x1 1 0.0297 0.0135 4.8535 0.0276 x2 1 0.4088 0.5990 0.4657 0.4950 x3 1 -0.3051 0.6041 0.2551 0.6135 x4 1 1.5746 0.5016 9.8543 0.0017
Odds Ratio Estimates
Point 95% Wald Effect Estimate Confidence Limits
x1 1.030 1.003 1.058 x2 1.505 0.465 4.868 x3 0.737 0.226 2.408 x4 4.829 1.807 12.907
Association of Predicted Probabilities and Observed Responses
Percent Concordant 77.5 Somers’ D 0.554 Percent Discordant 22.1 Gamma 0.556 Percent Tied 0.3 Tau-a 0.242 Pairs 2077 c 0.777
Classification Table
Correct Incorrect Percentages Prob Non- Non- Sensi- Speci- False False Level Event Event Event Event Correct tivity ficity POS NEG
0.060 31 0 67 0 31.6 100.0 0.0 68.4 . 0.080 31 4 63 0 35.7 100.0 6.0 67.0 0.0 0.100 29 12 55 2 41.8 93.5 17.9 65.5 14.3 0.120 29 22 45 2 52.0 93.5 32.8 60.8 8.3 0.140 28 23 44 3 52.0 90.3 34.3 61.1 11.5 0.160 27 25 42 4 53.1 87.1 37.3 60.9 13.8 0.180 26 32 35 5 59.2 83.9 47.8 57.4 13.5 0.200 26 36 31 5 63.3 83.9 53.7 54.4 12.2 0.220 25 39 28 6 65.3 80.6 58.2 52.8 13.3 0.240 23 41 26 8 65.3 74.2 61.2 53.1 16.3 0.260 22 42 25 9 65.3 71.0 62.7 53.2 17.6 0.280 20 43 24 11 64.3 64.5 64.2 54.5 20.4 0.300 20 45 22 11 66.3 64.5 67.2 52.4 19.6 0.320 19 46 21 12 66.3 61.3 68.7 52.5 20.7 0.340 18 48 19 13 67.3 58.1 71.6 51.4 21.3
The LOGISTIC Procedure
Classification Table
Correct Incorrect Percentages Prob Non- Non- Sensi- Speci- False False Level Event Event Event Event Correct tivity ficity POS NEG
0.360 17 50 17 14 68.4 54.8 74.6 50.0 21.9 0.380 16 51 16 15 68.4 51.6 76.1 50.0 22.7 0.400 14 51 16 17 66.3 45.2 76.1 53.3 25.0 0.420 13 53 14 18 67.3 41.9 79.1 51.9 25.4 0.440 13 53 14 18 67.3 41.9 79.1 51.9 25.4 0.460 12 53 14 19 66.3 38.7 79.1 53.8 26.4 0.480 12 55 12 19 68.4 38.7 82.1 50.0 25.7 0.500 11 55 12 20 67.3 35.5 82.1 52.2 26.7 0.520 10 56 11 21 67.3 32.3 83.6 52.4 27.3 0.540 10 58 9 21 69.4 32.3 86.6 47.4 26.6 0.560 9 59 8 22 69.4 29.0 88.1 47.1 27.2 0.580 8 61 6 23 70.4 25.8 91.0 42.9 27.4 0.600 8 62 5 23 71.4 25.8 92.5 38.5 27.1 0.620 8 62 5 23 71.4 25.8 92.5 38.5 27.1 0.640 7 64 3 24 72.4 22.6 95.5 30.0 27.3 0.660 7 64 3 24 72.4 22.6 95.5 30.0 27.3 0.680 6 64 3 25 71.4 19.4 95.5 33.3 28.1 0.700 5 64 3 26 70.4 16.1 95.5 37.5 28.9 0.720 5 65 2 26 71.4 16.1 97.0 28.6 28.6 0.740 5 65 2 26 71.4 16.1 97.0 28.6 28.6 0.760 3 65 2 28 69.4 9.7 97.0 40.0 30.1 0.780 2 65 2 29 68.4 6.5 97.0 50.0 30.9 0.800 1 67 0 30 69.4 3.2 100.0 0.0 30.9 0.820 1 67 0 30 69.4 3.2 100.0 0.0 30.9 0.840 0 67 0 31 68.4 0.0 100.0 . 31.6
Inputting the validation data set which is the remaining data from Data Set C.3, p. 1370.
data validation;
input id x1 socio x4 y x5;
label id = 'case'
x1 = 'age'
socio = 'socioeconomic status'
x4 = 'sector'
y = 'Disease status'
x5 = 'savings';
cards;
99 16 1 1 0 0
100 1 1 1 0 1
101 6 1 1 0 1
102 27 1 1 0 1
103 25 1 1 0 1
104 18 1 1 0 0
105 37 3 1 0 0
106 33 3 1 1 0
107 27 2 1 0 0
108 2 1 1 0 0
109 8 2 1 0 0
110 5 1 1 0 0
111 1 1 1 0 1
112 32 1 1 0 0
113 25 1 1 1 1
114 15 1 2 0 0
115 15 1 2 0 1
116 26 1 2 0 1
117 42 1 2 1 1
118 7 1 2 0 1
119 2 1 2 0 0
120 65 1 2 1 1
121 33 2 2 0 1
122 8 2 2 1 0
123 30 2 2 0 0
124 5 3 2 0 0
125 15 3 2 0 0
126 60 3 2 1 1
127 13 3 2 1 1
128 70 3 1 0 1
129 5 3 1 0 0
130 3 3 1 0 1
131 50 2 1 0 1
132 6 2 1 0 0
133 12 2 1 0 1
134 39 3 2 1 0
135 15 2 2 0 1
136 35 2 2 1 0
137 2 2 2 0 1
138 17 3 2 0 0
139 43 3 2 1 1
140 30 2 2 0 1
141 11 1 2 0 1
142 39 1 2 1 1
143 32 1 2 0 1
144 17 1 2 0 1
145 3 3 2 0 1
146 7 3 2 0 0
147 2 2 2 0 0
148 64 2 2 1 1
149 13 1 2 1 2
150 15 2 2 1 1
151 48 2 2 0 1
152 23 1 2 0 1
153 48 1 2 1 0
154 25 1 2 0 1
155 12 1 2 0 1
156 46 1 2 1 1
157 79 1 2 0 1
158 56 1 2 0 1
159 8 1 2 0 1
160 29 3 1 1 0
161 35 3 1 1 0
162 11 3 1 1 0
163 69 3 1 0 1
164 21 3 1 1 0
165 13 3 1 0 0
166 21 1 1 0 1
167 32 1 1 1 1
168 24 1 1 1 0
169 24 1 1 0 1
170 73 1 1 0 1
171 42 1 1 0 1
172 34 1 1 1 1
173 30 2 1 0 0
174 7 2 1 0 0
175 29 3 1 1 0
176 22 3 1 1 0
177 38 2 1 0 1
178 13 2 1 0 1
179 12 2 1 0 1
180 42 3 1 0 0
181 17 3 1 1 0
182 21 3 1 0 1
183 34 1 1 0 1
184 1 3 1 0 0
185 14 2 1 0 0
186 16 2 1 0 0
187 9 3 1 0 0
188 53 3 1 0 0
189 27 3 1 0 0
190 15 3 1 0 0
191 9 3 1 0 0
192 4 2 1 0 1
193 10 3 1 0 1
194 31 3 1 0 0
195 85 3 1 0 1
196 24 2 1 0 0
;
run;
Creating the dummy variables for the socioeconomic variable.
data validation; set validation; x2 = 0; if socio = 2 then x2 = 1; x3 = 0; if socio = 3 then x3 = 1; run;
Creating the fitted values of the validation dataset using parameter estimates from the Disease Outbreak dataset (table 14.3), p. 608. In order to get the same classification table it was necessary to use 0.7 as the cutoff value. The percentages shown in the table in the book are the column percentages which are in the second row of each cell.
Note: The proc format is simply to create nice labels for our table.
data validation1; set validation; e = 2.3129 - 0.0297*x1 - .4088*x2 + 0.3051*x3 - 1.5746*x4; ex = exp(e); p = 1/( 1+ ex); yes = 0; if p >= .7 then yes = 1; run; proc format; value y 1='with disease' 0='without disease'; value yes 1='pihat >= .7' 0='piehat < .7'; run; proc freq data = validation1; format y y. yes yes.; table yes*y / missing norow nopercent; run;
The FREQ ProcedureTable of yes by y
yes y(Disease status)
Frequency | Col Pct |without |with dis| Total |disease |ease | ————+——–+——–+ piehat < .7 | 44 | 12 | 56 | 61.11 | 46.15 | ————+——–+——–+ pihat >= .7 | 28 | 14 | 42 | 38.89 | 53.85 | ————+——–+——–+ Total 72 26 98
Inputting the Miller Lumber Company Example, p. 613.
data ch14tab08;
input y x1 x2 x3 x4 x5;
label x1 = 'Housing'
x2 = 'Income'
x3 = 'Age'
x4 = 'Competitor Distance'
x5 = 'Store Distance'
y = 'Costumers';
cards;
9 606 41393 3 3.04 6.32
6 641 23635 18 1.95 8.89
28 505 55475 27 6.54 2.05
11 866 64646 31 1.67 5.81
4 599 31972 7 0.72 8.11
4 520 41755 23 2.24 6.81
0 354 46014 26 0.77 9.27
14 483 34626 1 3.51 7.92
16 1034 85207 13 4.23 4.40
13 456 33021 32 3.07 6.03
9 19 39198 22 2.96 6.09
14 530 38794 5 2.77 6.08
5 337 30855 1 1.33 9.86
9 586 28852 7 2.98 8.64
9 1113 120065 9 3.58 5.26
7 525 32229 3 1.27 7.56
4 377 36828 15 1.92 8.91
26 1127 90302 26 5.83 1.74
32 877 51707 27 5.19 3.66
26 1007 89860 55 5.03 2.03
11 657 60513 32 4.38 8.30
12 302 42191 54 3.41 5.21
3 603 28736 41 0.34 8.29
15 556 49129 33 4.78 3.89
12 635 29308 42 2.53 6.17
9 386 26734 14 4.99 9.70
14 1011 57862 54 4.60 3.94
10 925 70030 36 4.58 8.66
22 898 46027 44 3.03 5.60
8 731 32202 43 5.15 9.67
3 584 32871 13 1.47 8.02
11 439 29564 18 3.67 5.10
2 153 46806 21 0.84 9.18
6 1069 59805 22 2.50 9.43
11 443 42555 53 2.62 5.75
10 392 36998 7 1.03 7.74
0 828 85664 4 1.30 9.66
15 159 21238 4 2.98 8.66
9 830 47972 40 2.28 9.26
16 234 33246 26 3.95 4.61
29 1004 45927 24 4.90 2.69
6 643 58315 8 0.78 6.26
26 741 69177 9 6.61 0.87
13 306 40886 27 4.53 2.68
0 180 44588 14 0.88 9.38
8 644 47347 35 2.94 7.69
8 109 31791 9 4.37 9.31
21 809 42740 17 4.10 4.75
12 722 59175 35 2.38 5.09
26 1006 48862 48 5.04 2.21
3 786 54678 20 3.59 8.52
7 1041 59835 40 1.68 7.59
5 524 51756 39 0.57 9.10
9 725 34817 18 1.88 7.96
13 482 29942 14 3.17 6.91
28 666 68684 25 5.78 2.55
10 450 64790 3 4.35 6.03
12 667 58535 25 2.78 5.59
6 921 42919 13 2.48 7.69
11 412 40722 32 2.47 9.43
12 526 42120 30 4.29 6.15
11 523 28647 43 2.69 7.54
9 1066 61464 40 1.15 8.25
8 1001 70136 29 2.58 9.67
9 669 34595 38 4.06 8.78
8 582 30878 58 1.91 6.86
6 872 39366 52 0.73 8.67
6 758 61563 31 3.08 8.33
15 782 38412 26 2.72 6.71
15 551 41045 2 3.62 7.45
12 201 23864 43 4.80 8.74
10 730 38647 9 0.67 7.92
8 738 58387 13 2.01 6.60
3 469 37242 40 1.42 8.37
10 898 38337 32 2.63 9.56
10 780 68201 5 4.12 6.69
15 622 41066 46 4.48 4.10
6 391 40873 19 1.67 6.90
9 531 54655 40 2.32 5.69
21 566 49826 1 3.06 4.03
13 410 29013 50 2.68 7.58
8 719 78082 31 2.70 4.89
6 684 57506 51 2.13 8.31
8 865 47118 46 2.17 9.06
21 1031 72373 48 6.27 1.75
7 862 67787 1 2.10 8.63
19 758 40305 15 3.95 5.58
13 1141 50026 45 2.79 6.18
24 1289 98701 8 5.87 2.73
7 674 58195 54 4.30 6.40
3 683 47991 57 1.54 9.52
8 650 63123 15 3.17 9.46
9 406 39051 29 3.11 9.62
18 966 114633 38 6.33 2.22
12 1103 55773 44 4.58 8.68
8 312 43393 41 2.25 6.43
16 787 61765 53 5.39 3.37
5 416 33348 48 1.48 7.66
8 528 44541 31 4.91 9.67
11 919 40795 8 2.97 7.79
12 482 55972 9 2.91 5.85
14 781 33140 30 1.42 5.71
17 120 19673 21 2.65 6.25
17 693 36190 6 4.70 9.54
6 348 25768 42 1.43 7.11
15 780 53974 47 4.21 6.41
10 752 71814 1 3.13 5.47
6 817 54429 47 1.90 9.90
4 268 34022 54 1.20 9.51
6 519 52850 43 2.92 8.62
;
run;
Table 14.9, p. 613.
Note: In SAS the estimate for beta2 -0.00001169 is rounded to -0.0000.
proc genmod data=ch14tab08; model y = x1-x5 / dist = poisson link = log; output out=temp p=muhati resdev=devi; run;
The GENMOD ProcedureModel Information
Data Set WORK.CH14TAB08 Distribution Poisson Link Function Log Dependent Variable y Costumers Observations Used 110
Criteria For Assessing Goodness Of Fit
Criterion DF Value Value/DF
Deviance 104 114.9854 1.1056 Scaled Deviance 104 114.9854 1.1056 Pearson Chi-Square 104 101.8808 0.9796 Scaled Pearson X2 104 101.8808 0.9796 Log Likelihood 1898.0224
Algorithm converged.
Analysis Of Parameter Estimates
Standard Wald 95% Confidence Chi- Parameter DF Estimate Error Limits Square Pr > ChiSq
Intercept 1 2.9424 0.2072 2.5362 3.3486 201.57 <.0001 x1 1 0.0006 0.0001 0.0003 0.0009 18.17 <.0001 x2 1 -0.0000 0.0000 -0.0000 -0.0000 30.63 <.0001 x3 1 -0.0037 0.0018 -0.0072 -0.0002 4.37 0.0365 x4 1 0.1684 0.0258 0.1179 0.2189 42.70 <.0001 x5 1 -0.1288 0.0162 -0.1605 -0.0970 63.17 <.0001 Scale 0 1.0000 0.0000 1.0000 1.0000
NOTE: The scale parameter was held fixed.
Table 14.10, p. 614.
proc print data = temp (obs=10); var y muhati devi; run;
Obs y muhati devi1 9 12.3378 -0.99880 2 6 8.7671 -0.99158 3 28 28.1259 -0.02375 4 11 8.4071 0.85335 5 4 7.2606 -1.32357 6 4 8.8818 -1.83900 7 0 4.2982 -2.93195 8 14 10.9989 0.86785 9 16 14.4440 0.40238 10 13 11.6344 0.39289
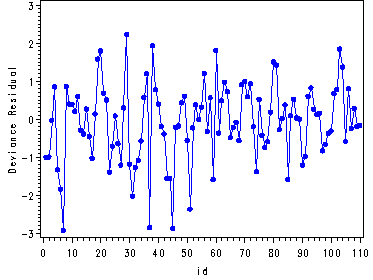
Fig. 14.9, p. 614.
Note: It is necessary to first create an index variable and graph the devi versus the index.
data temp; set temp; id = _n_; run; symbol1 v=dot i=join c=blue h = .8; axis1 label=(angle = 90); proc gplot data = temp; plot devi*id/ vaxis = axis1; run; quit;