In this chapter we will be using the hmohiv and the uis data sets.
Fig. 2.1, p. 28.
The first five observations of the hmohiv data set.
data mini; set hmohiv; if id > 5 then delete; run; proc print data=mini; var id time censor; run; Obs ID time censor 1 3 8 1 2 4 3 1 3 2 6 0 4 5 22 1 5 1 5 1
Table 2.2 and fig. 2.1, p. 32.
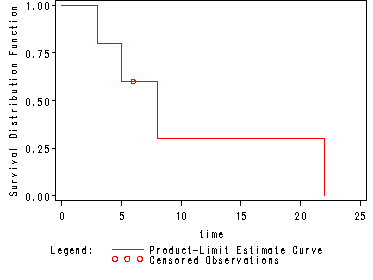
The estimated survivorship function computed for the mini data set in table 2.1, p. 28.
Note: The graph was generated by using the options plot=(s) where s=survival curve.
proc lifetest data=mini plots=(s);
time time*censor(0);
run;
<output omitted>
Product-Limit Survival Estimates
Survival
Standard Number Number
time Survival Failure Error Failed Left
0.0000 1.0000 0 0 0 5
3.0000 0.8000 0.2000 0.1789 1 4
5.0000 0.6000 0.4000 0.2191 2 3
6.0000* . . . 2 2
8.0000 0.3000 0.7000 0.2387 3 1
22.0000 0 1.0000 0 4 0
NOTE: The marked survival times are censored observations.
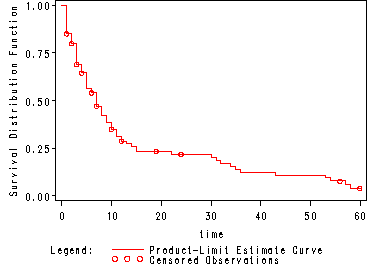
Fig. 2.2, p. 34.
The figure shows the Kaplan-Meier estimate of the survivorship function.
Note: We use the ods output system to create a data set est containing the Kaplan-Meier estimates and the components used in calculating the estimates.
ods listing close; ods output ProductLimitEstimates=est; proc lifetest data=hmohiv plots=(s); time time*censor(0); run; ods listing;
Table 2.3, p. 35.
Note: The output from proc lifetest has one line of output for each subject. In other words there would be 15 lines of output before the line containing the first estimate. We eliminate this extra output by including a where statement in the proc print stipulating that we do not wish to see the output where there is no Kaplan-Meier estimate.
proc print data=est noobs; where survival ne . ; var time left failed survival; run; <output omitted> time Left Failed Survival 0.0000 100 0 1.0000 1.0000 85 15 0.8500 2.0000 78 20 0.7988 3.0000 63 30 0.6894 4.0000 57 34 0.6442 5.0000 49 41 0.5636 ... 57.0000 3 79 0.0584 58.0000 2 80 0.0389
Creating the categorical variable for time.
data hmohivcat; set hmohiv; if time < 6 then timecat = 6; else if time < 12 then timecat = 12; else if time < 18 then timecat = 18; else if time < 24 then timecat = 24; else if time < 30 then timecat = 30; else if time < 36 then timecat = 36; else if time < 42 then timecat = 42; else if time < 48 then timecat = 48; else if time < 54 then timecat = 54; else if time < 60 then timecat = 60; else timecat = 66; run;
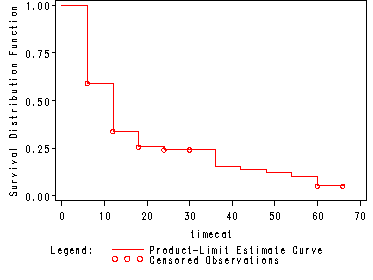
Fig. 2.3, p. 38.
Step function representation of life-table estimate of the survivorship function for the data in table 2.4.
proc lifetest data=hmohivcat plots=(s); time timecat*censor(0); run;
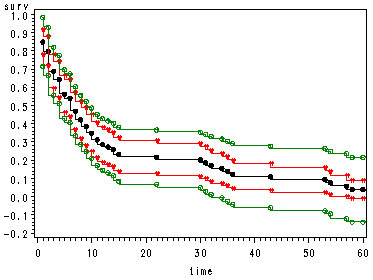
Fig. 2.5, p. 46.
This code is modified from SAS code used in the textbook example Survival Analysis by Klein and Moschberger.
This code was based on the formulas explained in section 4.4 of chapter 4, p. 100-103, and the confidence coefficient, k(aL, aU) = k(.145 .977) = 1.3581 at alpha = .05, was obtained in table C.4 of Appendix C.
The proc means is used to obtain the total number of observations (n=100) and the range of time, time = [0, 60]. The proc lifetest is used to obtain the values of the survival function estimate and the standard error of the survival function estimate at time=1 and time=60 which will be used to calculate sigma^2(1) and sigma^2(60) that are in turn used to calculate aL and aU.
proc means data=hmohiv; var time; run; proc lifetest data=hmohiv; time time*censor(0); run; proc sql; create table temp1 as select time, sum(censor) as num_event, count(time) as sub_total from hmohiv group by time; quit; data temp2; set temp1 nobs = all; total1 = lag(sub_total); retain num_left 100; /* total # of obs = 100 */ if _n_ > 1 then do; num_left = - total1 + num_left; end; retain surv 1 delta h sigma2 0; surv = surv*(1-num_event/num_left); delta= delta + num_event/(num_left*(num_left-num_event)); var =(surv**2)*delta; stderr = var**.5; h = h + num_event / num_left; sigma2 = sigma2 + num_event /(num_left)**2; stderrh = sigma2**.5; if num_event~=0 or _n_ = all; run; data temp3; set temp2; if surv ~=. then sigma = stderr / surv; md = probit(1-.05/2)*sigma ; md2 = 1.3581*(1+100*sigma2)/100**.5; /* 1.3581 = k(.145 .977) at alpha = .05*/ pointL = surv - md * surv; pointU = surv + md * surv; hwL = surv - md2*surv; hwU = surv + md2*surv; run; goptions reset=all; symbol1 value = dot h=.8 i = stepjr color = black; symbol2 value = ":" h=.8 i = stepjr color = red; symbol3 value = ":" h=.8 i = stepjr color = red; symbol4 value = circle h=.8 i = stepjr color = green; symbol5 value = circle h=.8 i = stepjr color = green; proc gplot data=temp3; plot (surv pointU pointL hwL hwU )*time /overlay; run; quit;
Graph legend:
Black = Kaplan-Meier estimate
Green = Point-wise 95% CI
Red = Hall & Wellner 95% CI.Table 2.5, p. 50.
ods output ProductLimitEstimates=est;
proc lifetest data=hmohiv;
time time*censor(0);
run;
<output omitted>
Quartile Estimates
Point 95% Confidence Interval
Percent Estimate [Lower Upper)
75 15.0000 10.0000 34.0000
50 7.0000 5.0000 9.0000
25 3.0000 2.0000 4.0000
Table 2.6, p. 52.
proc print data=est noobs;
where time ge 4 & time le 9 & survival ne .;
var time failed survival stderr;
run;
time Failed Survival StdErr
4.0000 34 0.6442 0.0493
5.0000 41 0.5636 0.0517
6.0000 43 0.5406 0.0521
7.0000 49 0.4701 0.0526
8.0000 53 0.4219 0.0525
9.0000 56 0.3857 0.0520
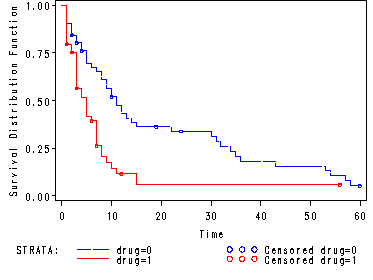
Fig. 2.7, p. 58.
Estimated survivorship functions for each group of drug use.
goptions reset = all; proc lifetest data=hmohiv plots=(s) noprint; time time*censor(0); strata drug; test age; run;
Table 2.8, p. 63.
Inputting the mini1 dataset.
data mini1; input time drug event; cards; 3 0 1 4 0 0 5 0 1 22 0 1 34 0 1 2 1 1 3 1 1 4 1 1 7 1 0 11 1 1 ; run; proc print data=mini1; run; time drug event 3 0 1 4 0 0 5 0 1 22 0 1 34 0 1 2 1 1 3 1 1 4 1 1 7 1 0 11 1 1
Table 2.10, p. 64.
Log-rank and Wilcoxon tests of equality of the survivorship functions across the two drug strata.
proc lifetest data=mini1;
time time*event(0);
strata drug;
run;
<output omitted>
Test of Equality over Strata
Pr >
Test Chi-Square DF Chi-Square
Log-Rank 1.5118 1 0.2189
Wilcoxon 1.2500 1 0.2636
-2Log(LR) 1.6489 1 0.1991
Table 2.11, p. 65.
Testing for equality of survivorship functions across drug strata for the hmohiv data.
proc lifetest data=hmohiv;
time time*censor(0);
strata drug;
run;
<output omitted>
Test of Equality over Strata
Pr >
Test Chi-Square DF Chi-Square
Log-Rank 11.8556 1 0.0006
Wilcoxon 10.9104 1 0.0010
-2Log(LR) 20.9264 1 <.0001
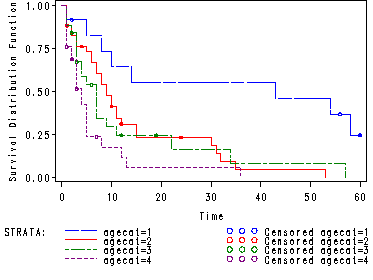
Creating the categorical variable for age.
data agecat; set hmohiv; if age le 29 then agecat = 1; else if age le 34 then agecat = 2; else if age le 39 then agecat = 3; else agecat = 4; run;
Table 2.12, p. 65.
Note: The median and 95% CIE for each stratum is found in the quartile section of the output for each stratum.
goptions reset=all;
proc lifetest data=agecat plots=(s);
time time*censor(0);
strata agecat;
symbol1 c=blue l=12 h=.5;
symbol2 c=red l=1 h=.5;
symbol3 c=green l=8 h=.5;
symbol4 c=purple l=2 h=.5;
run;
<output omitted>
Summary of the Number of Censored and Uncensored Values
Percent
Stratum agecat Total Failed Censored Censored
1 1 12 8 4 33.33
2 2 34 29 5 14.71
3 3 25 20 5 20.00
4 4 29 23 6 20.69
-------------------------------------------------------------------
Total 100 80 20 20.0
Quartile Estimates
Stratum 1: agecat = 1
Point 95% Confidence Interval
Percent Estimate [Lower Upper)
75 58.0000 43.0000 .
50 43.0000 8.0000 .
25 8.0000 5.0000 54.0000
Stratum 2: agecat = 2
Point 95% Confidence Interval
Percent Estimate [Lower Upper)
75 15.0000 10.0000 32.0000
50 9.0000 7.0000 12.0000
25 5.0000 2.0000 8.0000
Stratum 3: agecat = 3
Point 95% Confidence Interval
Percent Estimate [Lower Upper)
75 11.0000 7.0000 34.0000
50 7.0000 3.0000 9.0000
25 3.0000 2.0000 5.0000
Stratum 4: agecat = 4
Point 95% Confidence Interval
Percent Estimate [Lower Upper)
75 5.0000 4.0000 13.0000
50 4.0000 3.0000 5.0000
25 2.0000 1.0000 3.0000
Tables 2.14, page 70. The tests of the equality of the survivorship function across all strata.
Test of Equality over Strata
Pr >
Test Chi-Square DF Chi-Square
Log-Rank 19.9058 3 0.0002
Wilcoxon 14.1433 3 0.0027
-2Log(LR) 31.3597 3 <.0001
Fig. 2.8, p. 69.
Table 2.17, p. 76.
Note: SAS does not have any built in functionality which will compute the Nelson-Aalen estimates.
proc sql;
create table temp as
select time, sum(censor) as num_event, count(time) as sub_total
from hmohiv
group by time;
quit;
data temp1;
set temp;
total1 = lag(sub_total);
retain num_left 100; /* total obs = 100 */
if _n_ > 1 then do;
num_left = - total1 + num_left; end;
if num_event~=0;
retain surv 1 ;
surv = surv*(1-num_event/num_left);
keep time sub_total num_event num_left surv;
run;
/* Creating the Nelson-Aalen estimator (H tilda) and the Nelson-Aalen
estimator of the survivorship function (S tilda) */
data temp2;
set temp1;
retain h 0;
h = h + num_event/num_left;
hsurv = exp(-h);
run;
/* Renaming the sub_total to be called freq, num_event to be d and
num_left to be n in order to match the names used in the book. */
data temp3;
set temp2;
rename sub_total = freq;
rename num_event = d;
rename num_left = n;
run;
proc print data = temp3 noobs;
var time freq d n h hsurv surv;
run;
time freq d n h hsurv surv
1 17 15 100 0.15000 0.86071 0.85000
2 10 5 83 0.21024 0.81039 0.79880
3 12 10 73 0.34723 0.70664 0.68937
4 5 4 61 0.41280 0.66179 0.64417
5 7 7 56 0.53780 0.58403 0.56365
6 3 2 49 0.57862 0.56067 0.54064
7 7 6 46 0.70905 0.49211 0.47012
8 4 4 39 0.81162 0.44414 0.42190
9 3 3 35 0.89733 0.40766 0.38574
10 4 3 32 0.99108 0.37118 0.34958
11 3 3 28 1.09822 0.33346 0.31212
12 4 2 25 1.17822 0.30783 0.28715
13 1 1 21 1.22584 0.29351 0.27348
14 1 1 20 1.27584 0.27920 0.25981
15 2 2 19 1.38111 0.25130 0.23246
22 1 1 16 1.44361 0.23608 0.21793
30 1 1 14 1.51503 0.21980 0.20236
31 1 1 13 1.59196 0.20353 0.18680
32 1 1 12 1.67529 0.18725 0.17123
34 1 1 11 1.76620 0.17098 0.15566
35 1 1 10 1.86620 0.15471 0.14010
36 1 1 9 1.97731 0.13844 0.12453
43 1 1 8 2.10231 0.12217 0.10896
53 1 1 7 2.24517 0.10591 0.09340
54 1 1 6 2.41183 0.08965 0.07783
57 1 1 4 2.66183 0.06982 0.05837
58 1 1 3 2.99517 0.05003 0.03892
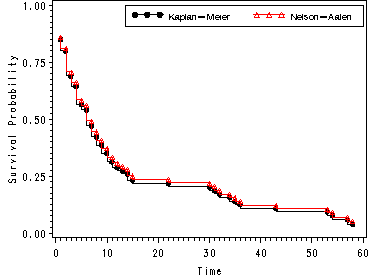
Fig. 2.10, p. 77.
Graphs of the Nelson-Aalen and Kaplan-Meier estimators of the survivorship function from the hmohiv data.
goptions reset=all;
symbol1 value = dot h=.8 i = stepjr color = black;
symbol2 value = triangle h=.8 i = stepjr color = red;
legend1 label=none value=(height=1 font=swiss 'Kaplan-Meier' 'Nelson-Aalen' )
position=(top right inside) mode=share cborder=black;
axis1 label=(a=90 'Survival Probability') order=(0 to 1.0 by .25);
proc gplot data=temp3;
plot (surv hsurv )*time /overlay legend=legend1 vaxis=axis1;
run;
quit;