The whas500, actg320, gbcs, uis and grace1000 data sets are used in this chapter.
Table 7.1 on page 211 using the actg320 data.
NOTE: The output from the proc means shown below is used in the data step to create the cd4 variable.
proc means data = actg320 p25 p50 p75; var cd4; run;
The MEANS Procedure
Analysis Variable : CD4
25th Pctl 50th Pctl 75th Pctl
--------------------------------------------
23.0000000 74.5000000 136.5000000
--------------------------------------------
data actg320_t71; set actg320; ivdrug_d = ivdrug>1; karnof_70=(karnof=70); karnof_80=(karnof=80); karnof_90=(karnof=90); karnof_70_80=(karnof=70 or karnof=80); * create stratified cd4 variable based on the output of the proc means above; cd4_q = 1; if cd4>23 & cd4<=74.5 then cd4_q=2; if cd4>74.5 & cd4<=136.5 then cd4_q=3; if cd4>136.5 then cd4_q=4; run; ods select FitStatistics ParameterEstimates; proc phreg data = actg320_t71; model time*censor(0) = tx ivdrug_d karnof_70_80 karnof_90 age; strata cd4_q; run;
Model Fit Statistics
Without With
Criterion Covariates Covariates
-2 LOG L 1050.245 1013.510
AIC 1050.245 1023.510
SBC 1050.245 1036.332
Analysis of Maximum Likelihood Estimates
Parameter Standard Hazard
Variable DF Estimate Error Chi-Square Pr > ChiSq Ratio
TX 1 -0.66777 0.21553 9.5991 0.0019 0.513
ivdrug_d 1 -0.54632 0.32256 2.8686 0.0903 0.579
karnof_70_80 1 1.19099 0.29630 16.1572 <.0001 3.290
karnof_90 1 0.41188 0.29267 1.9806 0.1593 1.510
AGE 1 0.02248 0.01120 4.0260 0.0448 1.023
Median values listed on page 211 using actg320 data.
data cov0; tx =0; ivdrug_d =0; karnof_70_80 =0; karnof_90 = 0; age = 0; run; proc phreg data = actg320_t71; model time*censor(0) = tx ivdrug_d karnof_70_80 karnof_90 age; strata cd4_q; output out=fig71_a xbeta=p; baseline covariates = cov0 out = b0 survival = surv / nomean; run; proc print data = b0; run; data t; set fig71_a; p1 = p - tx*(-0.66777); run; proc means data = t median; class cd4_q; var p1; output out=tcd4 (where = (_type_=1)) median = median; run; proc print data = tcd4; run; proc sql; select median into :md1 -:md4 from tcd4; quit;
median -------- 1.217544 1.176043 1.086145 1.093723
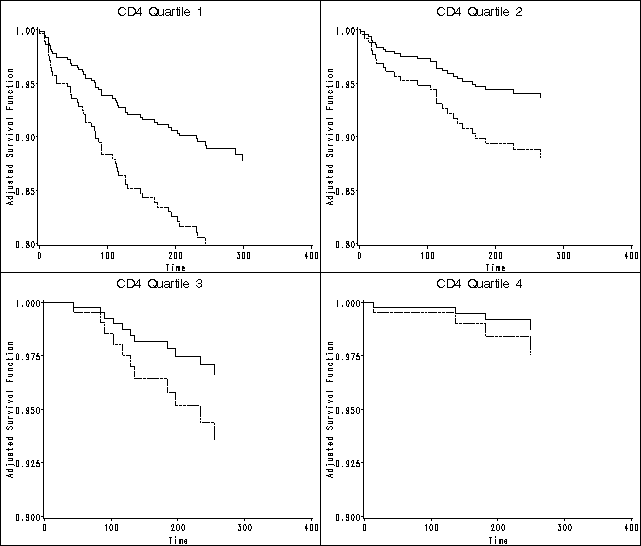
Figure 7.1 on page 212 continuing to use the actg320 data and the model in the previous example .
data fig71;
set b0;
s10 = surv**exp(&md1); /*cd4=1 and tr=0*/
s20 = surv**exp(&md2); /*cd4=2 and tr=0*/
s30 = surv**exp(&md3); /*cd4=3 and tr=0*/
s40 = surv**exp(&md4); /*cd4=4 and tr=0*/
s11 = surv**exp(&md1 -0.66777); /*cd4=1 and tr=1*/
s21 = surv**exp(&md2 -0.66777); /*cd4=2 and tr=1*/
s31 = surv**exp(&md3 -0.66777); /*cd4=3 and tr=1*/
s41 = surv**exp(&md4 -0.66777); /*cd4=4 and tr=1*/
run;
proc sort data = fig71;
by cd4_q time;
run;
goptions reset=all htext = 2 htitle = 3;
axis1 order=(.8 to 1 by .05) label=(a=90 'Adjusted Survival Function') minor=none;
axis2 order=(0 to 400 by 100) label=('Time') minor=none;
symbol1 i=stepjll v=none c=black line=14;
symbol2 i=stepjll v=none c=black line=1;
proc gplot data = fig71 ;
where cd4_q=1;
plot(s10 s11)*time /overlay vaxis=axis1 haxis=axis2 name="q1" noframe;
title "CD4 Quartile 1";
run;
quit;
goptions reset=all htext = 2 htitle = 3;
axis1 order=(.8 to 1 by .05) label=(a=90 'Adjusted Survival Function') minor=none;
axis2 order=(0 to 400 by 100) label=('Time') minor=none;
symbol1 i=stepjll v=none c=black line=14;
symbol2 i=stepjll v=none c=black line=1;
proc gplot data = fig71;
where cd4_q=2;
plot(s10 s11)*time /overlay vaxis=axis1 haxis=axis2 name="q2" noframe;
title "CD4 Quartile 2";
run;
quit;
goptions reset=all htext = 2 htitle = 3;
axis1 order=(.9 to 1 by .025) label=(a=90 'Adjusted Survival Function') minor=none;
axis2 order=(0 to 400 by 100) label=('Time') minor=none;
symbol1 i=stepjll v=none c=black line=14;
symbol2 i=stepjll v=none c=black line=1;
proc gplot data = fig71;
where cd4_q=3;
plot(s10 s11)*time /overlay vaxis=axis1 haxis=axis2 name="q3" noframe;
title "CD4 Quartile 3";
run;
quit;
* quantile 4 htext = 2 htitle = 3;
goptions reset=all htext = 2 htitle = 3;
axis1 order=(.9 to 1 by .025) label=(a=90 'Adjusted Survival Function') minor=none;
axis2 order=(0 to 400 by 100) label=('Time') minor=none;
symbol1 i=stepjll v=none c=black line=14;
symbol2 i=stepjll v=none c=black line=1;
proc gplot data = fig71;
where cd4_q=4;
plot(s10 s11)*time /overlay vaxis=axis1 haxis=axis2 name="q4" noframe;
title "CD4 Quartile 4";
run;
quit;
* empty graphics catalog;
proc greplay nofs igout=gseg;
delete _all_;
run;
quit;
proc greplay igout=work.gseg tc=sashelp.templt template=l2r2 nofs;
treplay 1:q1 2:q3 3:q2 4:q4;
run;
quit;
Estimates at the top of page 217 using the uis data.
ods select ParameterEstimates; proc phreg data = uis; model time*censor(0) = treat / risklimits; run;
The PHREG Procedure
Analysis of Maximum Likelihood Estimates
Parameter Standard
Variable DF Estimate Error Chi-Square Pr > ChiSq
treat 1 -0.23109 0.08899 6.7430 0.0094
Analysis of Maximum Likelihood Estimates
Hazard 95% Hazard Ratio
Variable Ratio Confidence Limits
treat 0.794 0.667 0.945
Table 7.2 on page 217 using the uis data.
ods output ParameterEstimates=out1; proc phreg data = uis; model time*censor(0) = treat off_trt tot; if time <= lot then off_trt = 0; else off_trt = 1; tot = treat*off_trt; run; data out2; set out1; z = estimate /stderr; lb = estimate-1.96*stderr; ub = estimate+1.96*stderr; run; proc print data = out2 noobs; var variable estimate stderr z probchisq lb ub; format z f8.2; format estimate probchisq lb ub f8.3; format stderr f8.4; run;
ProbChi Variable Estimate StdErr z Sq lb ub treat -0.524 0.2258 -2.32 0.020 -0.967 -0.081 off_trt 2.270 0.1865 12.17 0.000 1.905 2.636 tot 0.621 0.2463 2.52 0.012 0.138 1.104
Table 7.3 on page 219 using the uis data.
/* Lines 1 and 3 of table */ ods select ParameterEstimates; proc phreg data = uis; model time*censor(0) = treat off_trt tot / risklimits; if time <= lot then off_trt = 0; else off_trt = 1; tot = treat*off_trt; run;
The PHREG Procedure
Analysis of Maximum Likelihood Estimates
Parameter Standard
Variable DF Estimate Error Chi-Square Pr > ChiSq
treat 1 -0.52388 0.22583 5.3814 0.0204
off_trt 1 2.27033 0.18650 148.1847 <.0001
tot 1 0.62097 0.24630 6.3563 0.0117
Analysis of Maximum Likelihood Estimates
Hazard 95% Hazard Ratio
Variable Ratio Confidence Limits
treat 0.592 0.380 0.922
off_trt 9.683 6.718 13.955
tot 1.861 1.148 3.015
/* Line 2 of table */
ods select ParameterEstimates;
proc phreg data = uis;
model time*censor(0) = treat off_trt tot / risklimits;
if time <= lot then off_trt = 1;
else off_trt = 0;
tot = treat*off_trt;
run;
The PHREG Procedure
Analysis of Maximum Likelihood Estimates
Parameter Standard
Variable DF Estimate Error Chi-Square Pr > ChiSq
treat 1 0.09709 0.09771 0.9873 0.3204
off_trt 1 -2.27033 0.18650 148.1847 <.0001
tot 1 -0.62097 0.24630 6.3563 0.0117
Analysis of Maximum Likelihood Estimates
Hazard 95% Hazard Ratio
Variable Ratio Confidence Limits
treat 1.102 0.910 1.335
off_trt 0.103 0.072 0.149
tot 0.537 0.332 0.871
/* Line 4 of table */
ods select ParameterEstimates;
proc phreg data = uis;
model time*censor(0) = treat2 off_trt tot / risklimits;
treat2 = 1 - treat;
if time <= lot then off_trt = 0;
else off_trt = 1;
tot = treat2*off_trt;
run;
The PHREG Procedure
Analysis of Maximum Likelihood Estimates
Parameter Standard
Variable DF Estimate Error Chi-Square Pr > ChiSq
treat2 1 0.52388 0.22583 5.3814 0.0204
off_trt 1 2.89130 0.20501 198.8960 <.0001
tot 1 -0.62097 0.24630 6.3563 0.0117
Analysis of Maximum Likelihood Estimates
Hazard 95% Hazard Ratio
Variable Ratio Confidence Limits
treat2 1.689 1.085 2.629
off_trt 18.017 12.055 26.927
tot 0.537 0.332 0.871
Table 7.5 on page 222 using the grace1000 data.
data grace1000; set grace1000; age_inv = (1/age)*1000; sysbp_sqrt = sqrt(sysbp); run; ods output ParameterEstimates=out1; proc phreg data = grace1000; model days*death(0) = revasc age_inv sysbp_sqrt stchange; run; data out2; set out1; z = estimate /stderr; lb = estimate-1.96*stderr; ub = estimate+1.96*stderr; run; proc print data = out2 noobs; var variable estimate stderr z probchisq lb ub; format z f8.2; format estimate probchisq lb ub f8.3; format stderr f8.4; run;
ProbChi Variable Estimate StdErr z Sq lb ub REVASC -0.530 0.1172 -4.52 0.000 -0.759 -0.300 age_inv -0.189 0.0225 -8.41 0.000 -0.233 -0.145 sysbp_sqrt -0.207 0.0387 -5.36 0.000 -0.283 -0.132 STCHANGE 0.513 0.1188 4.32 0.000 0.280 0.746
Table 7.6 on page 223 using the grace1000 data.
ods output ParameterEstimates=out1;
proc phreg data = grace1000;
model days*death(0) = revasc_t age_inv sysbp_sqrt stchange;
if days <= revascdays or revasc = 0 then revasc_t = 0;
if days > revascdays and revasc = 1 then revasc_t = 1;
run;
data out2;
set out1;
z = estimate /stderr;
lb = estimate-1.96*stderr;
ub = estimate+1.96*stderr;
run;
proc print data = out2 noobs;
var variable estimate stderr z probchisq lb ub;
format z f8.2;
format estimate probchisq lb ub f8.3;
format stderr f8.4;
run;
ProbChi
Variable Estimate StdErr z Sq lb ub
revasc_t -0.261 0.1237 -2.11 0.035 -0.503 -0.019
age_inv -0.202 0.0228 -8.84 0.000 -0.247 -0.157
sysbp_sqrt -0.215 0.0392 -5.48 0.000 -0.292 -0.138
STCHANGE 0.501 0.1191 4.20 0.000 0.267 0.734
Table 7.7 on page 223 using the grace1000 data.
proc freq data = grace1000; tables revascdays; where revasc = 1; run;
The FREQ Procedure
Cumulative Cumulative
REVASCDAYS Frequency Percent Frequency Percent
---------------------------------------------------------------
0 151 28.93 151 28.93
1 81 15.52 232 44.44
2 38 7.28 270 51.72
3 30 5.75 300 57.47
4 17 3.26 317 60.73
5 13 2.49 330 63.22
6 11 2.11 341 65.33
7 14 2.68 355 68.01
8 37 7.09 392 75.10
9 33 6.32 425 81.42
10 31 5.94 456 87.36
11 22 4.21 478 91.57
12 17 3.26 495 94.83
13 21 4.02 516 98.85
14 6 1.15 522 100.00
Table 7.8, page 225 using the grace1000 data.
NOTE: This code does not reproduce the results shown in the text.
ods select ParameterEstimates; proc phreg data = grace1000; model days*death(0) = r_t0 r_t1 r_t2_3 r_t_4_7 r_t_8_10 r_t_11_14 age_inv sysbp_sqrt stchange; if days > revascdays and revascdays = 0 and revasc = 1 then r_t0 = 1; else r_t0 = 0; if days > revascdays and revascdays = 1 and revasc = 1 then r_t1 = 1; else r_t1 = 0; if days > revascdays and 2 <= revascdays <= 3 and revasc = 1 then r_t2_3 = 1; else r_t2_3 = 0; if days > revascdays and 4 <= revascdays <= 7 and revasc = 1 then r_t_4_7 = 1; else r_t_4_7 = 0; if days > revascdays and 8 <= revascdays <= 10 and revasc = 1 then r_t_8_10 = 1; else r_t_8_10 = 0; if days > revascdays and 11 <= revascdays <= 14 and revasc = 1 then r_t_11_14 = 1; else r_t_11_14 = 0; run;
The PHREG Procedure
Analysis of Maximum Likelihood Estimates
Parameter Standard Hazard
Variable DF Estimate Error Chi-Square Pr > ChiSq Ratio
r_t0 1 -0.47098 0.18238 6.6689 0.0098 0.624
r_t1 1 -0.69605 0.32685 4.5351 0.0332 0.499
r_t2_3 1 0.03345 0.26459 0.0160 0.8994 1.034
r_t_4_7 1 0.04626 0.29155 0.0252 0.8739 1.047
r_t_8_10 1 -0.08362 0.23808 0.1234 0.7254 0.920
r_t_11_14 1 0.05509 0.26910 0.0419 0.8378 1.057
age_inv 1 -0.20073 0.02290 76.8588 <.0001 0.818
sysbp_sqrt 1 -0.21989 0.03929 31.3267 <.0001 0.803
STCHANGE 1 0.54015 0.12102 19.9208 <.0001 1.716
Table 7.9, page 225 using the grace1000 data.
NOTE: This code does not reproduce the results shown in the text.
ods select ParameterEstimates; proc phreg data = grace1000; model days*death(0) = r_t_0_1 r_t_2_14 age_inv sysbp_sqrt stchange; if days > revascdays and 0 <= revascdays <= 1 and revasc = 1 then r_t_0_1 = 1; else r_t_0_1 = 0; if days > revascdays and 2 <= revascdays <= 14 and revasc = 1 then r_t_2_14 = 1; else r_t_2_14 = 0; run;
The PHREG Procedure
Analysis of Maximum Likelihood Estimates
Parameter Standard Hazard
Variable DF Estimate Error Chi-Square Pr > ChiSq Ratio
r_t_0_1 1 -0.52332 0.16556 9.9911 0.0016 0.593
r_t_2_14 1 0.00495 0.15243 0.0011 0.9741 1.005
age_inv 1 -0.20124 0.02288 77.3821 <.0001 0.818
sysbp_sqrt 1 -0.22004 0.03915 31.5869 <.0001 0.802
STCHANGE 1 0.54755 0.12003 20.8104 <.0001 1.729
Table 7.11, page 226 using the grace1000 data.
NOTE: This code does not reproduce the results shown in the text.
ods select ParameterEstimates; proc phreg data = grace1000; model days*death(0) = revasc_t0_1 revasc_t2_14 age_inv sysbp_sqrt stchange / risklimits; if revasc = 0 or (revasc = 1 and revascdays >= 15) then do; revasc_t0_1 = 0; revasc_t2_14 = 0; end; if revasc = 1 and (revascdays <2) then do; revasc_t0_1 = 1; revasc_t2_14 = 0; end; if revasc = 1 and 2 <= revascdays <= 14 then do; revasc_t0_1 = 0; revasc_t2_14 = 1; end; run;
The PHREG Procedure
Analysis of Maximum Likelihood Estimates
Parameter Standard
Variable DF Estimate Error Chi-Square Pr > ChiSq
revasc_t0_1 1 -0.64091 0.16157 15.7353 <.0001
revasc_t2_14 1 -0.44869 0.13851 10.4937 0.0012
age_inv 1 -0.18861 0.02250 70.2870 <.0001
sysbp_sqrt 1 -0.20953 0.03874 29.2488 <.0001
STCHANGE 1 0.53129 0.11998 19.6079 <.0001
Analysis of Maximum Likelihood Estimates
Hazard 95% Hazard Ratio
Variable Ratio Confidence Limits
revasc_t0_1 0.527 0.384 0.723
revasc_t2_14 0.638 0.487 0.838
age_inv 0.828 0.792 0.865
sysbp_sqrt 0.811 0.752 0.875
STCHANGE 1.701 1.345 2.152
Table 7.13 (text has no Table 7.12, but two 7.13s), page 229 using the whas500 data.
data t713a; set whas500; bmifp1=(bmi/10)**2; bmifp2=(bmi/10)**3; ag = age*gender; run; ods output ParameterEstimates=out1; proc phreg data = t713a; model lenfol*fstat(0) = bmifp1 bmifp2 age hr diasbp gender chf ag; where dstat = 0 and lenfol > los and id ~=416; run; data out2; set out1; z = estimate /stderr; lb = estimate-1.96*stderr; ub = estimate+1.96*stderr; run; proc print data = out2 noobs; var variable estimate stderr z probchisq lb ub; format z f8.2; format estimate probchisq lb ub f8.3; format stderr f8.4; run; Variable Estimate StdErr z Sq lb ub bmifp1 -0.728 0.1936 -3.76 0.000 -1.108 -0.349 bmifp2 0.156 0.0438 3.55 0.000 0.070 0.241 AGE 0.067 0.0093 7.16 0.000 0.048 0.085 HR 0.014 0.0033 4.30 0.000 0.008 0.020 DIASBP -0.012 0.0039 -3.14 0.002 -0.020 -0.005 GENDER 2.498 1.0632 2.35 0.019 0.415 4.582 CHF 0.903 0.1626 5.55 0.000 0.584 1.221 ag -0.037 0.0134 -2.79 0.005 -0.064 -0.011
Table 7.13 on page 235 using the gbcs data.
data ch7t13b; set gbcs; months = 12*rectime/365.25; if 0 < months <= 12 then interv = 1; if 12 < months <= 24 then interv = 2; if 24 < months <= 36 then interv = 3; if 36 < months <= 48 then interv = 4; if 48 < months <= 60 then interv = 5; if 60 < months <= 72 then interv = 6; if months > 72 then interv = 7; run; proc freq data = ch7t13b; tables interv*censrec; run;
The FREQ Procedure
Table of interv by CENSREC
interv CENSREC
Frequency|
Percent |
Row Pct |
Col Pct | 0| 1| Total
---------+--------+--------+
1 | 28 | 56 | 84
| 4.08 | 8.16 | 12.24
| 33.33 | 66.67 |
| 7.24 | 18.73 |
---------+--------+--------+
2 | 35 | 109 | 144
| 5.10 | 15.89 | 20.99
| 24.31 | 75.69 |
| 9.04 | 36.45 |
---------+--------+--------+
3 | 68 | 59 | 127
| 9.91 | 8.60 | 18.51
| 53.54 | 46.46 |
| 17.57 | 19.73 |
---------+--------+--------+
4 | 64 | 39 | 103
| 9.33 | 5.69 | 15.01
| 62.14 | 37.86 |
| 16.54 | 13.04 |
---------+--------+--------+
5 | 85 | 22 | 107
| 12.39 | 3.21 | 15.60
| 79.44 | 20.56 |
| 21.96 | 7.36 |
---------+--------+--------+
6 | 74 | 11 | 85
| 10.79 | 1.60 | 12.39
| 87.06 | 12.94 |
| 19.12 | 3.68 |
---------+--------+--------+
7 | 33 | 3 | 36
| 4.81 | 0.44 | 5.25
| 91.67 | 8.33 |
| 8.53 | 1.00 |
---------+--------+--------+
Total 387 299 686
56.41 43.59 100.00
Table 7.14 on page 237 using the modified gbcs data from the previous example.
data ch7t14; set ch7t13b; z = 0; month = interv*12; do interval = 1 to interv; if interval = interv then z =censrec ; newid = 8; if id = 230 then newid = 1; if id = 621 then newid = 2; if id = 17 then newid = 3; if id = 321 then newid = 4; if id = 117 then newid = 5; if id = 288 then newid = 6; if id = 278 then newid = 7; output; end; run; proc sort data = ch7t14; by newid; run; proc print data = ch7t14 noobs; where id in(230, 621, 17, 321, 117, 288, 278); var id month interval censrec z age; run;
ID month interval CENSREC z AGE 230 12 1 1 1 46 621 12 1 0 0 65 17 24 1 1 0 62 17 24 2 1 1 62 321 24 1 0 0 43 321 24 2 0 0 43 117 36 1 1 0 65 117 36 2 1 0 65 117 36 3 1 1 65 288 36 1 0 0 57 288 36 2 0 0 57 288 36 3 0 0 57 278 60 1 1 0 70 278 60 2 1 0 70 278 60 3 1 0 70 278 60 4 1 0 70 278 60 5 1 1 70
Table 7.15 on page 237 using the modified gbcs data from a previous example.
* Recoding hormone from 1/2 to 0/1; data ch7t15; set ch7t14; hormone = hormone - 1; run; ods select ParameterEstimates; proc genmod data = ch7t15 desc; class interval; model z = age hormone size interval / noint link = cloglog dist = bin; run;
Analysis Of Parameter Estimates
Standard Wald 95% Confidence Wald
Parameter DF Estimate Error Limits Chi-Square Pr > ChiSq
Intercept 0 0.0000 0.0000 0.0000 0.0000 . .
AGE 1 0.0014 0.0061 -0.0107 0.0134 0.05 0.8235
HORMONE 1 -0.3576 0.1283 -0.6090 -0.1062 7.77 0.0053
SIZE 1 0.0147 0.0036 0.0077 0.0218 16.80 <.0001
interval 1 1 -2.8734 0.3712 -3.6009 -2.1459 59.92 <.0001
interval 2 1 -2.0087 0.3595 -2.7134 -1.3040 31.21 <.0001
interval 3 1 -2.3647 0.3695 -3.0890 -1.6405 40.95 <.0001
interval 4 1 -2.4392 0.3780 -3.1799 -1.6984 41.65 <.0001
interval 5 1 -2.6302 0.4023 -3.4187 -1.8417 42.74 <.0001
interval 6 1 -2.6680 0.4525 -3.5549 -1.7812 34.77 <.0001
interval 7 1 -2.7639 0.6781 -4.0930 -1.4349 16.61 <.0001
Scale 0 1.0000 0.0000 1.0000 1.0000
NOTE: The scale parameter was held fixed.
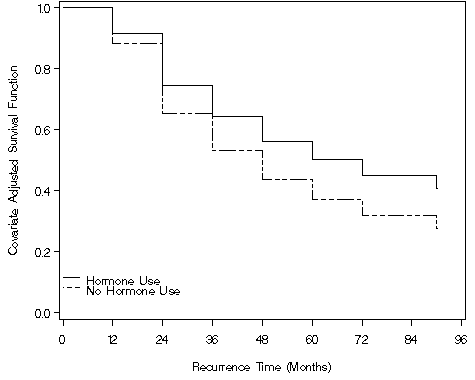
Figure 7.2, page 240
data ch7f2;
set ch7t14;
array dummy{7} int_1 - int_7;
do i = 1 to 7;
dummy(i) = 0;
end;
dummy(interval) = 1;
run;
ods output parameterestimates = b ;
proc genmod data = ch7f2 desc;
model z = age hormone size int_1 - int_7 / noint link = cloglog dist = bin;
run;
data b_1;
set b end=last;
retain age hormone size ;
retain s_h s_nh;
retain time 0;
if Parameter = "AGE" then age = estimate;
else if Parameter = "HORMONE" then hormone = estimate;
else if Parameter = "SIZE" then size = estimate;
else do;
if parameter ="int_1" then do;
s_nh = exp(-exp(estimate))**(exp(age*53 + size*25));
s_h = exp(-exp(estimate))**(exp(age*53 + size*25 + hormone));
time = 12;
end;
else if ~last then do;
s_nh = s_nh* exp(-exp(estimate))**(exp(age*53 + size*25));
s_h = s_h* exp(-exp(estimate))**(exp(age*53 + size*25 + hormone));
time = time + 12;
end;
end;
if parameter = "int_7" then time = 90;
if last then do;
time = 0;
s_nh = 1;
s_h = 1;
end;
run;
proc sort data = b_1;
by time;
run;
data annolegend;
length function style color $8 text $22;
retain xsys ysys '1' when 'a' color 'black';
function='move';
x=5; y=10; output;
function='draw';
x=1; y=10; line=14; output;
function='label';
x=7; y=10; position='6';
text='No Hormone Use';
output;
function='move';
x=5; y=13; output;
function='draw';
x=1; y=13; line=1; output;
function='label';
x=7; y=13; position='6';
text='Hormone Use';
output;
run;
goptions reset=all;
axis1 order=(0 to 1 by .2) label=(a=90 'Covariate Adjusted Survival Function') minor=none;
axis2 order=(0 to 96 by 12) label=('Recurrence Time (Months)') minor=none;
symbol1 i=stepjll v=none c=black line=1;
symbol2 i=stepjll v=none c=black line=14;
proc gplot data = b_1;
plot (s_h s_nh)*time /overlay vaxis=axis1 haxis = axis2 annotate = annolegend;
run;
quit;