Table 9.1 on page 155.
data table9_1;
input age $1-8 smoking $13-22 deaths person_years;
cards;
35 to 44 smoker 32 52407
45 to 54 smoker 104 43248
55 to 64 smoker 206 28612
65 to 74 smoker 186 12663
75 to 84 smoker 102 5317
35 to 44 non-smoker 2 18790
45 to 54 non-smoker 12 10673
55 to 64 non-smoker 28 5710
65 to 74 non-smoker 28 2585
75 to 84 non-smoker 31 1462
;
run;
options nodate pageno=1 linesize=105 pagesize=60;
proc tabulate data=table9_1 format=9.;
class age smoking /order=data;
var deaths person_years;
table age='age group'*sum='',
smoking=''*(deaths person_years )
/ RTS=20 row=float;
run;
------------------------------------------------------------ | | smoker | non-smoker | | |-------------------+-------------------| | | |person_y-| |person_y-| | | deaths | ears | deaths | ears | |------------------+---------+---------+---------+---------| |age group | | | | | |------------------| | | | | |35 to 44 | 32| 52407| 2| 18790| |------------------+---------+---------+---------+---------| |45 to 54 | 104| 43248| 12| 10673| |------------------+---------+---------+---------+---------| |55 to 64 | 206| 28612| 28| 5710| |------------------+---------+---------+---------+---------| |65 to 74 | 186| 12663| 28| 2585| |------------------+---------+---------+---------+---------| |75 to 84 | 102| 5317| 31| 1462| ------------------------------------------------------------
Figure 9.1 on page 155.
data fig9_1; set table9_1; y = deaths/person_years*100000; run; goptions reset = all; symbol1 v = dot c=black; symbol2 v = P font = marker c = blue; axis1 order = (0 to 2500 by 500) label=(a=90 'Deaths per 100,00 person years'); proc gplot data = fig9_1; plot y*age = smoking /vaxis=axis1; run; quit;
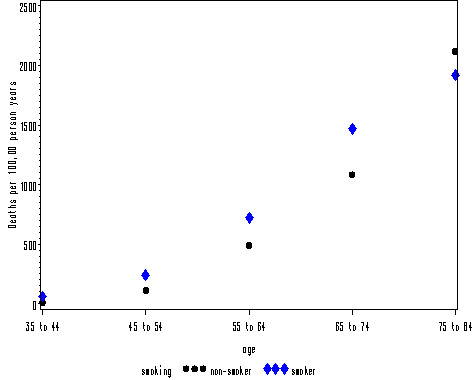
Table 9.2 on page 156.
data table9_2; set table9_1; logd = log(deaths); logp = log(person_years); if age = '35 to 44' then agecat = 1; if age = '45 to 54' then agecat = 2; if age = '55 to 64' then agecat = 3; if age = '65 to 74' then agecat = 4; if age = '75 to 84' then agecat = 5; agesq = agecat*agecat; smoke = (smoking='smoker'); smkage = agecat*smoke; run; proc genmod data = table9_2; model deaths = agecat agesq smoke smkage /d=poi offset = logp; run; quit;
Criteria For Assessing Goodness Of Fit
Criterion DF Value Value/DF
Deviance 5 1.6354 0.3271
Scaled Deviance 5 1.6354 0.3271
Pearson Chi-Square 5 1.5503 0.3101
Scaled Pearson X2 5 1.5503 0.3101
Log Likelihood 2727.6433
Analysis Of Parameter Estimates
Standard Wald 95% Confidence Chi-
Parameter DF Estimate Error Limits Square Pr > ChiSq
Intercept 1 -10.7918 0.4501 -11.6739 -9.9096 574.92 <.0001
agecat 1 2.3765 0.2079 1.9689 2.7841 130.60 <.0001
agesq 1 -0.1977 0.0274 -0.2513 -0.1440 52.17 <.0001
smoke 1 1.4410 0.3722 0.7115 2.1705 14.99 0.0001
smkage 1 -0.3075 0.0970 -0.4977 -0.1174 10.04 0.0015
Scale 0 1.0000 0.0000 1.0000 1.0000
Table 9.3 on page 157.
proc genmod data = table9_2; model deaths = agecat agesq smoke smkage /obstats d=poi offset = logp; ods output obstats = table9_3; run; quit; proc print data = table9_3; var agecat smoke deaths pred reschi resdev; run;
Obs agecat smoke deaths Pred Reschi Resdev 1 1 1 32 29.584734 0.4440493 0.438204 2 2 1 104 106.81196 -0.272082 -0.273289 3 3 1 206 208.19865 -0.152376 -0.152645 4 4 1 186 182.82789 0.2345992 0.2339257 5 5 1 102 102.57677 -0.056948 -0.057001 6 1 0 2 3.4148015 -0.765619 -0.83049 7 2 0 12 11.541629 0.1349222 0.1340436 8 3 0 28 24.743377 0.6546934 0.6410667 9 4 0 28 30.229155 -0.405441 -0.410583 10 5 0 31 31.071038 -0.012744 -0.012749
Table 9.4 on page 157.
data table9_4;
input type $1-30 site $32-44 frequency;
cards;
hutchinson's melanotic freckle head & neck 22
hutchinson's melanotic freckle trunk 2
hutchinson's melanotic freckle extremities 10
superficial spreading melanoma head & neck 16
superficial spreading melanoma trunk 54
superficial spreading melanoma extremities 115
nodular head & neck 19
nodular trunk 33
nodular extremities 73
indeterminate head & neck 11
indeterminate trunk 17
indeterminate extremities 28
;
run;
options nodate pageno=1 linesize=105 pagesize=60;
proc tabulate data=table9_4 format=9.;
class type site /order=data;
var frequency;
table ( type='Tumor type' all='Total')*sum='',
(site='Site' all='Total')*(frequency='')
/ RTS=20 row=float;
run;
------------------------------------------------------------ | | Site | | | |-----------------------------| | | | head & | |extremit-| | | | neck | trunk | ies | Total | |------------------+---------+---------+---------+---------| |Tumor type | | | | | |------------------| | | | | |hutchinson's | | | | | |melanotic freckle | 22| 2| 10| 34| |------------------+---------+---------+---------+---------| |superficial | | | | | |spreading melanoma| 16| 54| 115| 185| |------------------+---------+---------+---------+---------| |nodular | 19| 33| 73| 125| |------------------+---------+---------+---------+---------| |indeterminate | 11| 17| 28| 56| |------------------+---------+---------+---------+---------| |Total | 68| 106| 226| 400| ------------------------------------------------------------
Table 9.5 on page 158.
options nodate pageno=1 linesize=105 pagesize=60;
proc tabulate data=table9_4 ;
class type site /order=data;
var frequency;
table ( type='Tumor type' all='Total')*rowpctsum='',
(site='Site' all='Total')*(frequency='')
/ RTS=20 row=float;
run;
------------------------------------------------------------------------
| | Site | |
| |--------------------------------------| |
| |head & neck | trunk |extremities | Total |
|------------------+------------+------------+------------+------------|
|Tumor type | | | | |
|------------------| | | | |
|hutchinson's | | | | |
|melanotic freckle | 64.71| 5.88| 29.41| 100.00|
|------------------+------------+------------+------------+------------|
|superficial | | | | |
|spreading melanoma| 8.65| 29.19| 62.16| 100.00|
|------------------+------------+------------+------------+------------|
|nodular | 15.20| 26.40| 58.40| 100.00|
|------------------+------------+------------+------------+------------|
|indeterminate | 19.64| 30.36| 50.00| 100.00|
|------------------+------------+------------+------------+------------|
|Total | 17.00| 26.50| 56.50| 100.00|
------------------------------------------------------------------------
proc tabulate data=table9_4 ;
class type site /order=data;
var frequency;
table ( type='Tumor type' all='Total')*colpctsum='',
(site='Site' all='Total')*(frequency='')
/ RTS=20 row=float;
run;
------------------------------------------------------------------------ | | Site | | | |--------------------------------------| | | |head & neck | trunk |extremities | Total | |------------------+------------+------------+------------+------------| |Tumor type | | | | | |------------------| | | | | |hutchinson's | | | | | |melanotic freckle | 32.35| 1.89| 4.42| 8.50| |------------------+------------+------------+------------+------------| |superficial | | | | | |spreading melanoma| 23.53| 50.94| 50.88| 46.25| |------------------+------------+------------+------------+------------| |nodular | 27.94| 31.13| 32.30| 31.25| |------------------+------------+------------+------------+------------| |indeterminate | 16.18| 16.04| 12.39| 14.00| |------------------+------------+------------+------------+------------| |Total | 100.00| 100.00| 100.00| 100.00| ------------------------------------------------------------------------
Table 9.6 on page 159.
data table9_6;
input treatment $1-7 response $9-16 frequency;
cards;
placebo small 25
placebo moderate 8
placebo large 5
vaccine small 6
vaccine moderate 18
vaccine large 11
;
run;
options nodate pageno=1 linesize=105 pagesize=60;
proc tabulate data=table9_6 format=9.;
class treatment response /order=data;
var frequency;
table ( treatment='' )*sum='',
(response='Response' all='Total')*(frequency='')
/ RTS=20 row=float;
run;
------------------------------------------------------------ | | Response | | | |-----------------------------| | | | small |moderate | large | Total | |------------------+---------+---------+---------+---------| |placebo | 25| 8| 5| 38| |------------------+---------+---------+---------+---------| |vaccine | 6| 18| 11| 35| ------------------------------------------------------------
Table 9.7 on page 160.
data table9_7;
length ulcer $10;
input ulcer $ case_control $ aspirin $ frequency;
cards;
gastric control non-user 62
gastric control user 6
gastric case non-user 39
gastric case user 25
duodenal control non-user 53
duodenal control user 8
duodenal case non-user 49
duodenal case user 8
;
run;
options nodate pageno=1 linesize=105 pagesize=60;
proc tabulate data=table9_7 format=9.;
class ulcer case_control aspirin /order=data;
var frequency;
table (ulcer='' )*case_control=''*sum='',
(aspirin='Aspirin use' all='Total')*(frequency='')
/ RTS=20 row=float;
run;
-------------------------------------------------- | | Aspirin use | | | |-------------------| | | |non-user | user | Total | |------------------+---------+---------+---------| |gastric |control | 62| 6| 68| | |---------+---------+---------+---------| | |case | 39| 25| 64| |--------+---------+---------+---------+---------| |duodenal|control | 53| 8| 61| | |---------+---------+---------+---------| | |case | 49| 8| 57| --------------------------------------------------
Table 9.8 on page 160.
options nodate pageno=1 linesize=105 pagesize=60;
proc tabulate data=table9_7 format=9.;
class ulcer case_control aspirin /order=data;
var frequency;
table (ulcer='' )*case_control=''*rowpctsum='',
(aspirin='Aspirin use' all='Total')*(frequency='')
/ RTS=20 row=float;
run;
-------------------------------------------------- | | Aspirin use | | | |-------------------| | | |non-user | user | Total | |------------------+---------+---------+---------| |gastric |control | 91| 9| 100| | |---------+---------+---------+---------| | |case | 61| 39| 100| |--------+---------+---------+---------+---------| |duodenal|control | 87| 13| 100| | |---------+---------+---------+---------| | |case | 86| 14| 100| --------------------------------------------------
Table 9.9 on page 165.
proc freq data = table9_4 order=data; weight frequency; tables type*site /chisq expected norow nocol nopercent; run;
type site
Frequency |
Expected |head & n|trunk |extremit| Total
|eck | |ies |
-----------------+--------+--------+--------+
hutchinson's mel | 22 | 2 | 10 | 34
anotic freckle | 5.78 | 9.01 | 19.21 |
-----------------+--------+--------+--------+
superficial spre | 16 | 54 | 115 | 185
ading melanoma | 31.45 | 49.025 | 104.53 |
-----------------+--------+--------+--------+
nodular | 19 | 33 | 73 | 125
| 21.25 | 33.125 | 70.625 |
-----------------+--------+--------+--------+
indeterminate | 11 | 17 | 28 | 56
| 9.52 | 14.84 | 31.64 |
-----------------+--------+--------+--------+
Total 68 106 226 400
Statistics for Table of type by site
Statistic DF Value Prob
------------------------------------------------------
Chi-Square 6 65.8129 <.0001
Likelihood Ratio Chi-Square 6 51.7950 <.0001
Mantel-Haenszel Chi-Square 1 2.4161 0.1201
Phi Coefficient 0.4056
Contingency Coefficient 0.3759
Cramer's V 0.2868
Sample Size = 400
Table 9.10 on page 166.
Saturated model (9.10).
data table9_4a; set table9_4; if type = "hutchinson's melanotic freckle" then ntype = 3; if type = "superficial spreading melanoma" then ntype = 2; if type = "nodular" then ntype = 1; if type = "indeterminate" then ntype = 0; if site = 'head & neck' then nsite = 2; if site = 'trunk' then nsite = 1; if site = 'extremities' then nsite = 0; run; proc genmod data = table9_4a order=internal ; class ntype nsite; model frequency = ntype|nsite@2 /d=poi; run;
Analysis Of Parameter Estimates
Standard Wald 95% Confidence Chi-
Parameter DF Estimate Error Limits Square Pr > ChiSq
Intercept 1 3.0910 0.2132 2.6732 3.5089 210.20 <.0001
ntype 0 1 -0.6931 0.3693 -1.4169 0.0306 3.52 0.0605
ntype 1 1 -0.1466 0.3132 -0.7604 0.4672 0.22 0.6397
ntype 2 1 -0.3185 0.3286 -0.9624 0.3255 0.94 0.3324
ntype 3 0 0.0000 0.0000 0.0000 0.0000 . .
nsite 0 1 -0.7885 0.3814 -1.5360 -0.0410 4.27 0.0387
nsite 1 1 -2.3979 0.7385 -3.8454 -0.9504 10.54 0.0012
nsite 2 0 0.0000 0.0000 0.0000 0.0000 . .
ntype*nsite 0 0 1 1.7228 0.5216 0.7004 2.7451 10.91 0.0010
ntype*nsite 0 1 1 2.8332 0.8338 1.1990 4.4674 11.55 0.0007
ntype*nsite 0 2 0 0.0000 0.0000 0.0000 0.0000 . .
ntype*nsite 1 0 1 2.1345 0.4602 1.2325 3.0365 21.51 <.0001
ntype*nsite 1 1 1 2.9500 0.7927 1.3963 4.5036 13.85 0.0002
ntype*nsite 1 2 0 0.0000 0.0000 0.0000 0.0000 . .
ntype*nsite 2 0 1 2.7608 0.4655 1.8485 3.6731 35.18 <.0001
ntype*nsite 2 1 1 3.6143 0.7915 2.0630 5.1656 20.85 <.0001
ntype*nsite 2 2 0 0.0000 0.0000 0.0000 0.0000 . .
ntype*nsite 3 0 0 0.0000 0.0000 0.0000 0.0000 . .
ntype*nsite 3 1 0 0.0000 0.0000 0.0000 0.0000 . .
ntype*nsite 3 2 0 0.0000 0.0000 0.0000 0.0000 . .
Scale 0 1.0000 0.0000 1.0000 1.0000
Additive model (9.9).
proc genmod data = table9_4a order=internal ; class ntype nsite; model frequency = ntype nsite /d=poi; run;
Criteria For Assessing Goodness Of Fit
Criterion DF Value Value/DF
Deviance 6 51.7950 8.6325
Scaled Deviance 6 51.7950 8.6325
Pearson Chi-Square 6 65.8129 10.9688
Scaled Pearson X2 6 65.8129 10.9688
Log Likelihood 1124.3272
Analysis Of Parameter Estimates
Standard Wald 95% Confidence Chi-
Parameter DF Estimate Error Limits Square Pr > ChiSq
Intercept 1 1.7544 0.2040 1.3546 2.1542 73.96 <.0001
ntype 0 1 0.4990 0.2174 0.0729 0.9251 5.27 0.0217
ntype 1 1 1.3020 0.1934 0.9229 1.6811 45.31 <.0001
ntype 2 1 1.6940 0.1866 1.3283 2.0597 82.42 <.0001
ntype 3 0 0.0000 0.0000 0.0000 0.0000 . .
nsite 0 1 1.2010 0.1383 0.9299 1.4721 75.40 <.0001
nsite 1 1 0.4439 0.1554 0.1394 0.7485 8.16 0.0043
nsite 2 0 0.0000 0.0000 0.0000 0.0000 . .
Scale 0 1.0000 0.0000 1.0000 1.0000
Minimal model.
proc genmod data = table9_4a ; model frequency = /d=poi; run;
Criteria For Assessing Goodness Of Fit
Criterion DF Value Value/DF
Deviance 11 295.2030 26.8366
Scaled Deviance 11 295.2030 26.8366
Pearson Chi-Square 11 348.7394 31.7036
Scaled Pearson X2 11 348.7394 31.7036
Log Likelihood 1002.6232
Algorithm converged.
Analysis Of Parameter Estimates
Standard Wald 95% Confidence Chi-
Parameter DF Estimate Error Limits Square Pr > ChiSq
Intercept 1 3.5066 0.0500 3.4086 3.6046 4918.39 <.0001
Scale 0 1.0000 0.0000 1.0000 1.0000
Table 9.11 on page 167. SAS has different formula for the likelihood function. But the difference of the maximum likelihood of two models is the same as shown in the book. In the following code, we used ods listing close option to temporarily turn off displaying output in the output window. The ods output statement creates a data file contains the model fit information. After all the models are run, we concatenate all the model fit information into one single file. We can do the same comparison described at the end of page 166. For example, the deviance difference for the second and third model is 2*(669.1102 – 663.4848) = 11.2508. The deviance difference for the third model and the fourth is 2*(671.2379 – 669.1102) = 4.2554. They are the same as shown in the book.
ods listing close; proc genmod data = table9_7; class ulcer case_control ; model frequency = ulcer|case_control@2 / d=poisson; ods output modelfit = m1; run; data m1; set m1; retain mdf; if _n_= 1 then mdf = df; m = 'm1'; if _n_ = 5; keep m mdf value; run; proc genmod data = table9_7; class ulcer case_control aspirin; model frequency = ulcer|case_control@2 aspirin / d=poisson; ods output modelfit = m2; run; data m2; set m2; retain mdf; if _n_= 1 then mdf = df; m = 'm2'; if _n_ = 5; keep m mdf value; run; proc genmod data = table9_7; class ulcer case_control aspirin; model frequency = ulcer|case_control@2 aspirin aspirin*case_control / d=poisson; ods output modelfit = m3; run; data m3; set m3; retain mdf; if _n_= 1 then mdf = df; m = 'm3'; if _n_ = 5; keep m mdf value; run; proc genmod data = table9_7; class ulcer case_control aspirin; model frequency = ulcer|case_control|aspirin@2 / d=poisson; ods output modelfit = m4; run; data m4; set m4; retain mdf; if _n_= 1 then mdf = df; m = 'm4'; if _n_ = 5; keep m mdf value; run; data all; set m1 m2 m3 m4; run; ods listing; proc print data = all; run;
Obs Value mdf m 1 611.0255 4 m1 2 663.4848 3 m2 3 669.1102 2 m3 4 671.2379 1 m4
Table 9.12 on page 167.
proc genmod data = table9_7; class ulcer case_control aspirin; model frequency = ulcer|case_control|aspirin@2 /obstats d=poisson; ods output obstats = table9_12; run;
Criteria For Assessing Goodness Of Fit
Criterion DF Value Value/DF
Deviance 1 6.2830 6.2830
Scaled Deviance 1 6.2830 6.2830
Pearson Chi-Square 1 6.4880 6.4880
Scaled Pearson X2 1 6.4880 6.4880
Log Likelihood 671.2379
proc tabulate data=table9_12 ;
class ulcer case_control aspirin /order=data;
var frequency pred;
table (ulcer='' )*case_control=''*sum='',
(aspirin='Aspirin use' all='Total')*(frequency='' pred='')
/ RTS=20 row=float;
run;
-------------------------------------------------------------------------------------------------- | | Aspirin use | | | |---------------------------------------------------| | | | non-user | user | Total | |------------------+-------------------------+-------------------------+-------------------------| |gastric |control | 62.00| 58.53| 6.00| 9.47| 68.00| 68.00| | |---------+------------+------------+------------+------------+------------+------------| | |case | 39.00| 42.47| 25.00| 21.53| 64.00| 64.00| |--------+---------+------------+------------+------------+------------+------------+------------| |duodenal|control | 53.00| 56.47| 8.00| 4.53| 61.00| 61.00| | |---------+------------+------------+------------+------------+------------+------------| | |case | 49.00| 45.53| 8.00| 11.47| 57.00| 57.00| --------------------------------------------------------------------------------------------------
