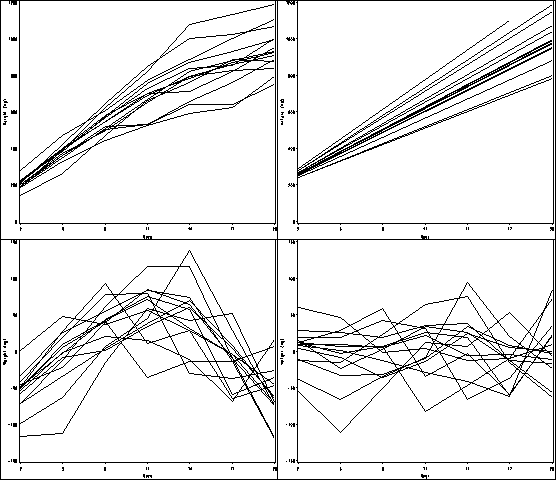
Figure 10.1, page 334.
*Figure a; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(2 to 20 by 3) label=(a=0 'Days') minor=none; axis2 order =(0 to 1200 by 200) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = smallmice; plot weight*day=id / haxis = axis1 vaxis = axis2 nolegend name = 'f10_1a'; run; quit; *Figure b; data sm; set smallmice; cont_day = day; run; proc mixed data = sm method = reml; class id day; model weight = cont_day/ outpred = pred_sm; random cont_day/ subject = id type = un; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(2 to 20 by 3) label=(a=0 'Days') minor=none; axis2 order =(000 to 1200 by 200) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = pred_sm; plot pred*day=id / haxis = axis1 vaxis = axis2 nolegend name = 'f10_1b'; run; quit; *Figure c; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(2 to 20 by 3) label=(a=0 'Days') minor=none; axis2 order =(-150 to 150 by 50) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = pred_sm; plot resid*day=id / haxis = axis1 vaxis = axis2 nolegend name = 'f10_1c'; run; quit; *Figure d; proc mixed data = sm method = reml; class id day; model weight = cont_day cont_day*cont_day/ outpred = pred_sm2; random cont_day/ subject = id type = un; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(2 to 20 by 3) label=(a=0 'Days') minor=none; axis2 order =(-150 to 150 by 50) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = pred_sm2; plot resid*day=id / haxis = axis1 vaxis = axis2 nolegend name = 'f10_1d'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=l2r2 nofs; treplay 1:f10_1a 2:f10_1c 3:f10_1b 4:f10_1d; run; quit;
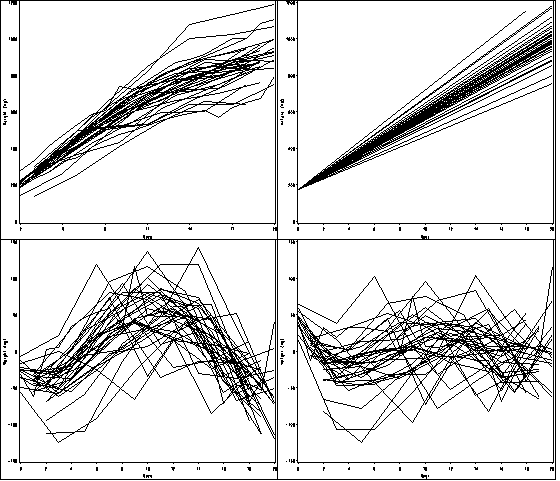
Figure 10.2, page 335.
*Figure a; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(0 to 20 by 2) label=(a=0 'Days') minor=none; axis2 order =(0 to 1200 by 200) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = bigmice; plot weight*day=id / haxis = axis1 vaxis = axis2 nolegend name = 'f10_2a'; run; quit; *Figure b; data bm; set bigmice; cont_day = day; run; proc mixed data = bm method = reml; class id day; model weight = cont_day/ outpred = pred_bm; random cont_day/ subject = id type = un; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(0 to 20 by 2) label=(a=0 'Days') minor=none; axis2 order =(000 to 1200 by 200) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = pred_bm; plot pred*day=id / haxis = axis1 vaxis = axis2 nolegend name = 'f10_2b'; run; quit; *Figure c; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(0 to 20 by 2) label=(a=0 'Days') minor=none; axis2 order =(-150 to 150 by 50) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = pred_bm; plot resid*day=id / haxis = axis1 vaxis = axis2 nolegend name = 'f10_2c'; run; quit; *Figure d; proc mixed data = bm method = reml; class id day; model weight = cont_day cont_day*cont_day/ outpred = pred_bm2; random cont_day/ subject = id type = un; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(0 to 20 by 2) label=(a=0 'Days') minor=none; axis2 order =(-150 to 150 by 50) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = pred_bm2; plot resid*day=id / haxis = axis1 vaxis = axis2 nolegend name = 'f10_2d'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=l2r2 nofs; treplay 1:f10_2a 2:f10_2c 3:f10_2b 4:f10_2d; run; quit;
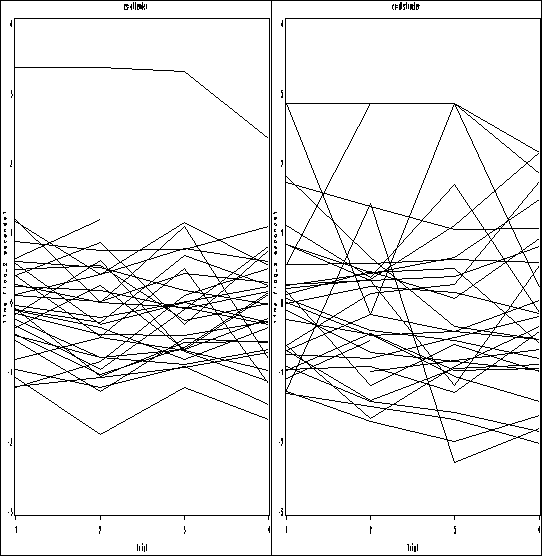
Figure 10.3, page 337.
data pain1; set pain; tmt_a = (treatment = "attend")*(trial = 4); tmt_d = (treatment = "distract")*(trial = 4); tmt_n = (treatment = "no directions")*(trial = 4); attend = (cs = "attender"); distract = (cs = "distracter"); run; proc mixed data = pain1 method = reml; class id trial; model l2paintol = attend tmt_a tmt_d tmt_n attend*tmt_a attend*tmt_d attend*tmt_n/ outpred=pred_pain; repeated trial / subject = id type = cs; run; proc sort data = pred_pain; by cs id trial; run; proc greplay igout = work.gseg nofs; delete _all_; run; quit; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 100; axis1 order =(1 to 4 by 1) label=(a=0 'Trial') minor=none; axis2 order =(-3 to 4 by 1) label = (a=90 'Time (log_2 seconds)') minor=none ; proc gplot data = pred_pain; by cs; plot resid*trial=id/ nolegend haxis=axis1 vaxis=axis2 name='f10_3_'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=h2 nofs; treplay 1:f10_3_ 2:f10_3_1 ; run; quit;
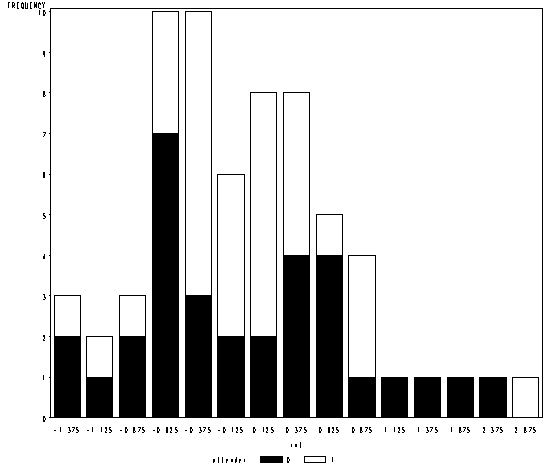
Figure 10.4, page 338.
proc mixed data = pain1 method = reml noclprint noitprint noinfo; class id trial; model l2paintol = attend tmt_a tmt_d tmt_n attend*tmt_a attend*tmt_d attend*tmt_n/ notest outpred = id_est; random intercept / subject = id type = un solution; run; data est_intercept; input id estimate @@; datalines; 1 -0.3043 2 -0.6488 3 -1.0046 4 -0.6417 5 0.1990 6 1.2456 7 -0.4754 8 -0.8621 9 0.1353 10 0.04389 11 -1.4032 12 0.1224 13 -0.2475 14 0.5574 15 -0.6272 16 0.3819 17 0.9213 18 0.02049 19 -0.7358 20 -0.4957 21 2.8233 22 0.6626 23 0.3961 24 0.3321 25 -1.3005 26 0.01034 27 -0.6336 28 -0.3554 29 -0.7229 30 -0.4311 31 1.3668 32 -0.8595 33 0.7519 34 0.7446 35 -1.4561 36 -0.1218 37 -0.3138 38 -0.01444 39 0.5920 40 -0.2831 41 -0.3140 42 0.8265 43 0.07633 44 -0.6576 45 -0.6538 46 -0.2161 47 -1.2259 48 -0.8144 49 0.1901 50 0.8077 51 -0.4188 52 0.7474 53 0.4312 54 -0.5192 55 -0.7178 56 2.4837 57 0.2918 58 0.4356 59 -0.4888 60 1.9018 61 0.3491 62 0.4075 63 -0.1313 64 -0.1592 ; run; proc sql; create table id_cs as select cs, mean(id) as id from id_est group by id, cs; quit; proc sort data = id_cs; by id; run; data intercept; merge id_cs est_intercept; by id; run; data intercept; set intercept; int = .; if (-1.5 <= estimate < -1.25) then int = -1.375; if (-1.25 <= estimate < -1) then int = -1.125; if (-1 <= estimate < -.75) then int = -.875; if (-.75 <= estimate < -.5) then int = -.625; if (-.5 <= estimate lt -.25) then int = -.375; if (-.25 <= estimate lt 0) then int = -.125; if (0 <= estimate lt .25) then int = .125; if (.25 <= estimate lt .5) then int = .375; if (.5 <= estimate lt .75) then int = .625; if (.75 <= estimate lt 1) then int = .875; if (1 <= estimate lt 1.25) then int = 1.125; if (1.25 <= estimate lt 1.5) then int = 1.375; if (1.5 <= estimate lt 1.75) then int = 1.625; if (1.75 <= estimate lt 2) then int = 1.875; if (2 <= estimate lt 2.25) then int = 2.125; if (2.25 <= estimate lt 2.5) then int = 2.375; if (2.5 <= estimate lt 2.75) then int = 2.625; if (2.75 <= estimate lt 3) then int = 2.875; attender = (cs = "attender"); run; goptions colors=(black white); proc gchart data = intercept; vbar int/subgroup=attender discrete minor =0;* midpoints=(-1.5 to 3 by .25); run; quit;
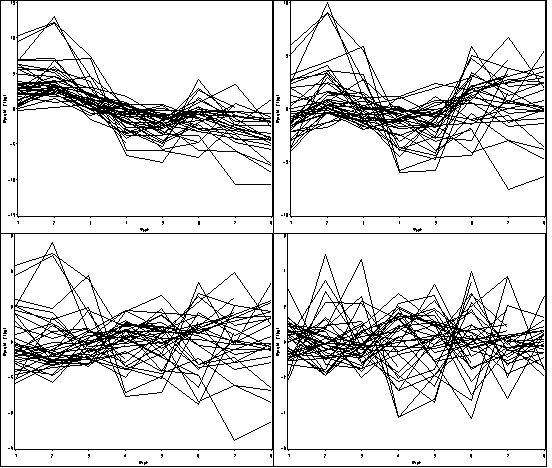
Figure 10.5, page 339.
proc greplay igout = work.gseg nofs; delete _all_; run; quit; *a; proc mixed data=weight1 method = reml; class id; model weight = / outpred=resid_f10_5a; random intercept/subject = id type = un; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(1 to 8 by 1) label=(a=0 'Week') minor=none; axis2 order =(-15 to 15 by 5) label = (a=90 'Weight (lbs)') minor=none ; proc gplot data = resid_f10_5a ; plot resid*week=id / haxis = axis1 vaxis = axis2 nolegend name='f10_5a'; run; quit; *b; proc mixed data=weight1 method = reml; class id; model weight = week / outpred=resid_f10_5b; random intercept/subject = id type = un; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(1 to 8 by 1) label=(a=0 'Week') minor=none; axis2 order =(-10 to 10 by 5) label = (a=90 'Weight (lbs)') minor=none ; proc gplot data = resid_f10_5b ; plot resid*week=id / haxis = axis1 vaxis = axis2 nolegend name='f10_5b'; run; quit; *c; proc mixed data=weight1 method = reml; class id week; model weight = week / outpred=resid_f10_5c; random intercept week/subject = id type = cs; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(1 to 8 by 1) label=(a=0 'Week') minor=none; axis2 order =(-9 to 9 by 3) label = (a=90 'Weight (lbs)') minor=none ; proc gplot data = resid_f10_5c ; plot resid*week=id / haxis = axis1 vaxis = axis2 nolegend name='f10_5c'; run; quit; *d; data weight1; set weight1; cont_week=week; run; proc mixed data=weight1 method = reml; class id week; model weight = week / outpred=resid_f10_5d; random intercept cont_week/subject = id type = un; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(1 to 8 by 1) label=(a=0 'Week') minor=none; axis2 order =(-6 to 6 by 2) label = (a=90 'Weight (lbs)') minor=none ; proc gplot data = resid_f10_5d; plot resid*week=id / haxis = axis1 vaxis = axis2 nolegend name='f10_5d'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=l2r2 nofs; treplay 1:f10_5a 2:f10_5c 3:f10_5b 4:f10_5d; run; quit;