The data files can be downloaded from
http://rem.ph.ucla.edu/~rob/mld/data.html .
Table 12.1, page 367.
*Complete data;
proc mixed data = missingdata1 method = reml noinfo noclprint noitprint;
class idno;
model weight = week/ solution notest;
random intercept/ subject = idno type = un;
run;
The Mixed Procedure
Covariance Parameter Estimates
Cov Parm Subject Estimate
UN(1,1) idno 714.01
Residual 10.1683
Fit Statistics
-2 Res Log Likelihood 735.4
AIC (smaller is better) 739.4
AICC (smaller is better) 739.5
BIC (smaller is better) 741.4
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
1 384.48 <.0001
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 196.46 5.9972 19 32.76 <.0001
week 0.1414 0.08522 99 1.66 0.1002
*MAR;
proc mixed data = missingdata2 method = reml noclprint noitprint noinfo;
class idno;
model weight = week/ solution notest;
random intercept/ subject = idno type =un;
run;
The Mixed Procedure
Covariance Parameter Estimates
Cov Parm Subject Estimate
UN(1,1) idno 700.56
Residual 7.9755
Fit Statistics
-2 Res Log Likelihood 560.5
AIC (smaller is better) 564.5
AICC (smaller is better) 564.6
BIC (smaller is better) 566.5
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
1 301.54 <.0001
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 196.84 5.9377 19 33.15 <.0001
week -0.05619 0.09671 70 -0.58 0.5631
*MNAR;
proc mixed data = missingdata3 method = reml noclprint noitprint noinfo;
class idno;
model weight = week/ solution notest;
random intercept/ subject = idno type =un;
run;
The Mixed Procedure
Covariance Parameter Estimates
Cov Parm Subject Estimate
UN(1,1) idno 707.88
Residual 6.0963
Fit Statistics
-2 Res Log Likelihood 490.2
AIC (smaller is better) 494.2
AICC (smaller is better) 494.4
BIC (smaller is better) 496.2
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
1 280.88 <.0001
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 196.29 5.9666 19 32.90 <.0001
week -0.2007 0.08905 60 -2.25 0.0279
Table 12.2, page 370.
proc sql;
create table schiz as
select *,
(max(week) ne 6) as drop_out,
sqrt(week) as sweek
from schizophrenia
group by id
order by drop_out, id;
quit;
*Pooled;
proc mixed data = schiz method = ml noitprint noclprint noinfo;
class id ;
model severity = sweek drug sweek*drug / solution notest;
random intercept sweek/ subject=id type = un;
run;
The Mixed Procedure
Covariance Parameter Estimates
Cov Parm Subject Estimate
UN(1,1) ID 0.3687
UN(2,1) ID 0.02085
UN(2,2) ID 0.2420
Residual 0.5778
Fit Statistics
-2 Log Likelihood 4649.0
AIC (smaller is better) 4665.0
AICC (smaller is better) 4665.1
BIC (smaller is better) 4697.6
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
3 542.43 <.0001
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 5.3480 0.08790 435 60.84 <.0001
sweek -0.3361 0.06794 435 -4.95 <.0001
drug 0.04634 0.1011 729 0.46 0.6469
sweek*drug -0.6405 0.07752 729 -8.26 <.0001
*By dropout;
proc mixed data = schiz method = ml noitprint noclprint noinfo;
by drop_out;
class id ;
model severity = sweek drug sweek*drug / solution notest;
random intercept sweek/ subject=id type = un;
run;
drop_out=0
The Mixed Procedure
Covariance Parameter Estimates
Cov Parm Subject Estimate
UN(1,1) ID 0.3984
UN(2,1) ID -0.01110
UN(2,2) ID 0.2048
Residual 0.5600
Fit Statistics
-2 Log Likelihood 3782.1
AIC (smaller is better) 3798.1
AICC (smaller is better) 3798.2
BIC (smaller is better) 3828.6
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
3 468.38 <.0001
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 5.2212 0.1091 333 47.84 <.0001
sweek -0.3934 0.07345 333 -5.36 <.0001
drug 0.2016 0.1226 655 1.64 0.1008
sweek*drug -0.5386 0.08255 655 -6.52 <.0001
drop_out=1
Covariance Parameter Estimates
Cov Parm Subject Estimate
UN(1,1) ID 0.2858
UN(2,1) ID 0.02045
UN(2,2) ID 0.6921
Residual 0.5147
Fit Statistics
-2 Log Likelihood 822.9
AIC (smaller is better) 838.9
AICC (smaller is better) 839.5
BIC (smaller is better) 859.9
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
3 73.32 <.0001
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 5.5337 0.1409 100 39.26 <.0001
sweek -0.1235 0.1773 100 -0.70 0.4877
drug -0.1939 0.1779 74 -1.09 0.2791
sweek*drug -1.1674 0.2239 74 -5.21 <.0001
*Covariate and contrasts;
proc mixed data = schiz method = ml noitprint noclprint noinfo;
class id ;
model severity = sweek drug sweek*drug drop_out drop_out*sweek
drop_out*drug drop_out*sweek*drug / solution notest;
random intercept sweek/ subject=id type = un;
estimate 'int placebo nodrop' intercept 1;
estimate 'int placebo drop' intercept 1 drop_out 1;
estimate 'int drug nodrop' intercept 1 drug 1;
estimate 'int drug drop' intercept 1 drug 1 drop_out 1;
estimate 'slope placebo nodrop' sweek 1;
estimate 'slope placebo drop' sweek 1 drop_out*sweek 1;
estimate 'slope drug nodrop' sweek 1 sweek*drug 1;
estimate 'slope drug drop' sweek 1 sweek*drug 1 drop_out*sweek 1 drop_out*sweek*drug 1;
run;
The Mixed Procedure
Covariance Parameter Estimates
Cov Parm Subject Estimate
UN(1,1) ID 0.3612
UN(2,1) ID 0.01175
UN(2,2) ID 0.2300
Residual 0.5768
Fit Statistics
-2 Log Likelihood 4623.3
AIC (smaller is better) 4647.3
AICC (smaller is better) 4647.5
BIC (smaller is better) 4696.2
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
3 523.49 <.0001
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 5.2210 0.1075 433 48.55 <.0001
sweek -0.3934 0.07635 433 -5.15 <.0001
drug 0.2017 0.1208 729 1.67 0.0955
sweek*drug -0.5386 0.08580 729 -6.28 <.0001
drop_out 0.3203 0.1864 729 1.72 0.0862
sweek*drop_out 0.2517 0.1593 729 1.58 0.1146
drug*drop_out -0.3987 0.2270 729 -1.76 0.0794
sweek*drug*drop_out -0.6348 0.1960 729 -3.24 0.0013
Estimates
Standard
Label Estimate Error DF t Value Pr > |t|
int placebo nodrop 5.2210 0.1075 433 48.55 <.0001
int placebo drop 5.5413 0.1523 729 36.39 <.0001
int drug nodrop 5.4227 0.05511 729 98.39 <.0001
int drug drop 5.7430 0.1944 729 29.54 <.0001
slope placebo nodrop -0.3934 0.07635 433 -5.15 <.0001
slope placebo drop -0.1417 0.1399 729 -1.01 0.3115
slope drug nodrop -0.9320 0.03916 729 -23.80 <.0001
slope drug drop -1.3150 0.1073 729 -12.26 <.0001
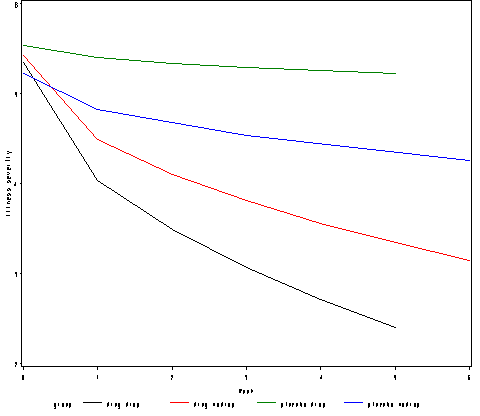
Figure 12.2, page 371.
proc mixed data = schiz method = ml noitprint noclprint noinfo; class id ; model severity = sweek drug sweek*drug drop_out drop_out*sweek drop_out*drug drop_out*sweek*drug / solution notest outpredm=pred_schiz;; random intercept sweek/ subject=id type = un; run; proc sql; create table pred_schiz2 as select mean(pred) as pred, drug, drop_out, week from pred_schiz group by week, drop_out, drug order by drop_out, drug, week; quit; data pred_schiz2; set pred_schiz2; group=compress(drug||drop_out); if group = "01" then group = "placebo drop"; if group = "00" then group = "placebo nodrop"; if group = "11" then group = "drug drop"; if group = "10" then group = "drug nodrop"; run; goptions reset = all; symbol1 value=none interpol=join r=4; axis1 order =(0 to 6 by 1 ) label=(a=0 'Week') minor=none; axis2 order =(2 to 6 by 1) label = (a=90 'Illness severity') minor=none ; proc gplot data = pred_schiz2; plot pred*week=group /haxis=axis1 vaxis = axis2; run; quit;