Figure 2.1, page 31.
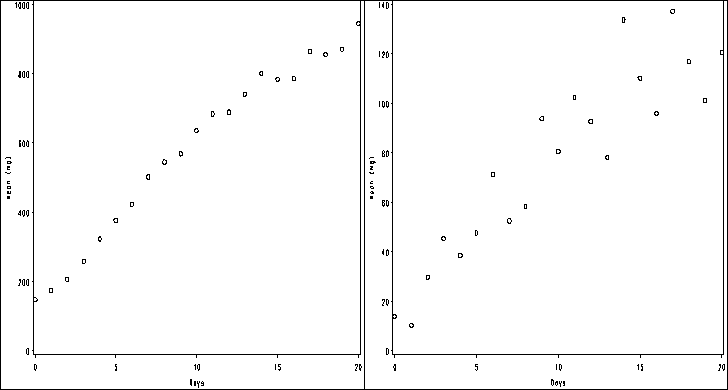
goptions reset = all; symbol1 value=circle color=black h=1; axis1 order =(0 to 20 by 5) label=(a=0 'Days') minor=none; axis2 order =(0 to 1200 by 200) label = (a=90 'Weight (mg)') minor=none; proc gplot data = bigmice; plot weight*day / haxis = axis1 vaxis = axis2; run;

Figure 2.2, page 32.
proc sort data = bigmice; by day; run; goptions reset = all; proc boxplot data = bigmice; plot weight*day / haxis=(0 to 20 by 5) vaxis = (0 to 1200 by 200) hminor=0 vminor=0; label weight = 'Weight'; label day = 'Day'; run;
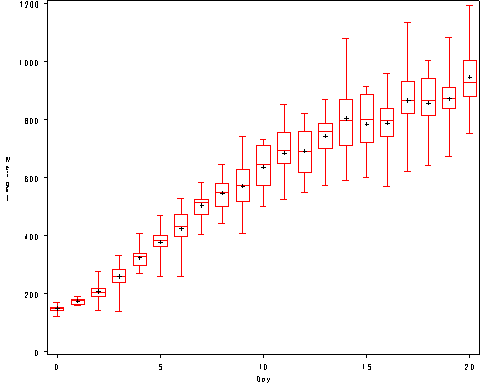
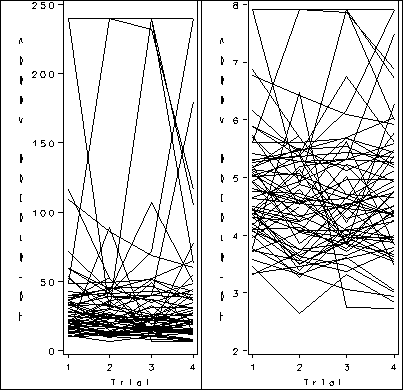
Figure 2.3, page 34.
proc sort data = bigmice; by day id; run; goptions reset = all; symbol1 value=circle color=black interpol = join repeat = 38; axis1 order =(0 to 20 by 5) label=(a=0 'Days') minor=none; axis2 order =(0 to 1200 by 200) label = (a=90 'Weight (mg)') minor=none ; proc gplot data = bigmice; plot weight*day=id / haxis = axis1 vaxis = axis2 nolegend; run; quit;
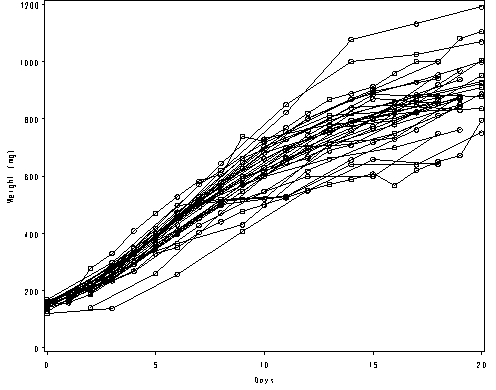
Figure 2.6, page 41.
proc sql;
create table summary as
select day,
mean(weight) as mean_wght,
std(weight) as sd_wght
from bigmice
group by day;
quit;
goptions reset = all;
symbol1 value=circle color=black;
axis1 order =(0 to 20 by 5) label=(a=0 'Days') minor=none;
axis2 label = (a=90 'mean (mg)') order=(0 to 1000 by 200) minor = none;
proc gplot data = summary; plot mean_wght*day / haxis=axis1 vaxis = axis2 name = 'f2_6a'; run; quit; goptions reset = all; symbol1 value=circle color=black; axis1 order =(0 to 20 by 5) label=(a=0 'Days') minor=none; axis2 order =(0 to 140 by 20) label = (a=90 'mean (mg)') minor = none; proc gplot data = summary; plot sd_wght*day / haxis=axis1 vaxis = axis2 name = 'f2_6b'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=h2 nofs; treplay 1:f2_6a 2:f2_6b; run; quit;
Figure 2.7, page 42.
proc sort data = pain; by day trial; run; goptions reset = all ; symbol1 value=none color=black interpol = join repeat = 100; axis1 order =(1 2 3 4) label=(a=0 'Trial') minor=none; axis2 order =(0 to 250 by 50) label = (a=90 'Tolerance (sec)') minor=none ; proc gplot data = pain; plot paintol*trial=id / haxis = axis1 vaxis = axis2 nolegend name = 'f2_7a'; run; quit; goptions reset = all ; symbol1 value=none color=black interpol = join repeat = 100; axis1 order =(1 2 3 4) label=(a=0 'Trial') minor=none; axis2 label = (a=90 'Tolerance (sec)') minor=none ; proc gplot data = pain; plot l2paintol*trial=id / haxis = axis1 vaxis = axis2 nolegend name = 'f2_7b'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=h2 nofs; treplay 1:f2_7a 2:f2_7b; run; quit;
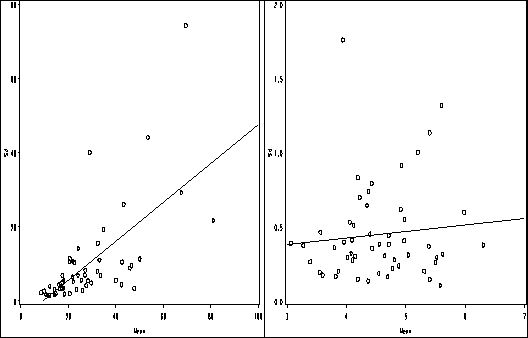
Figure 2.9, page 45.
proc sql;
create table pain0 as
select *,
max(paintol) as max_pain,
count(paintol) as count_paintol
from pain
group by id
having max(paintol) ne 240 and count(paintol) eq 4;
quit;
proc sql;
create table pain1 as
select id,
mean(paintol) as mean_pain,
std(paintol) as std_pain,
mean(l2paintol) as mean_l2pain,
std(l2paintol) as std_l2pain
from pain0
group by id;
quit;
goptions reset = all;
symbol1 value=circle color=black interpol = r;
axis1 order =(0 to 100 by 20) label=(a=0 'Mean') minor=none;
axis2 order =(0 to 80 by 20) label=(a=90 'Sd') minor = none;
proc gplot data = pain1;
plot std_pain*mean_pain / haxis=axis1 vaxis = axis2 name = 'f2_9a';
run; quit;
goptions reset = all;
symbol1 value=circle color=black interpol = r;
axis1 order =(3 to 7 by 1) label=(a=0 'Mean') minor=none;
axis2 order =(0 to 2 by .5) label=(a=90 'Sd') minor = none;
proc gplot data = pain1;
plot std_l2pain*mean_l2pain / haxis=axis1 vaxis = axis2 name = 'f2_9b';
run; quit;
proc greplay igout = work.gseg tc=sashelp.templt template=h2 nofs;
treplay 1:f2_9a 2:f2_9b;
run; quit;
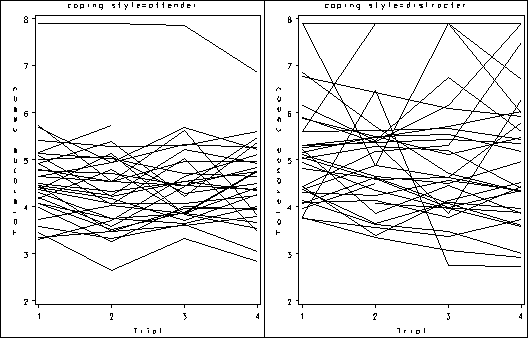
Figure 2.10, page 46.
proc sort data = pain; by cs id trial; run; goptions reset = all ; symbol1 value=none color=black interpol = join repeat = 100; axis1 order =(1 2 3 4) label=(a=0 'Trial') minor=none; axis2 label = (a=90 'Tolerance (sec)') minor=none ; proc gplot data = pain; by cs; plot l2paintol*trial=id / haxis = axis1 vaxis = axis2 nolegend name = 'f2_10_'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=h2 nofs; treplay 1:f2_10_ 2:f2_10_1; run; quit;
Table 2.1, page 48.
Note that there is no variable valley.
proc sql;
create table ozone as
select fullname, site,
mean(long) as long,
mean(lat) as lat,
mean(alt) as altitude
from newozone1
group by site, fullname;
quit;
proc print data = ozone noobs;
run;
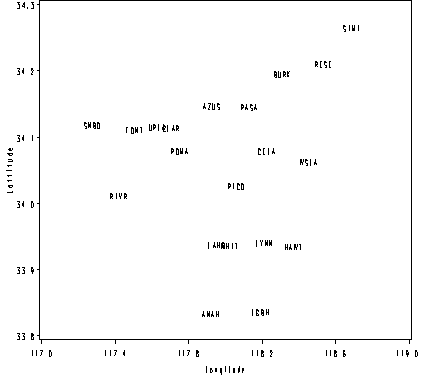
fullname site long lat altitude
ANAHEIM ANAH 117.919 33.821 41
AZUSA AZUS 117.923 34.136 189
BURBANK BURK 118.308 34.183 168
LOS-ANGELES-NORTH-MAIN CELA 118.225 34.067 87
CLAREMONT-COLLEGE CLAR 117.704 34.102 364
FONTANA-ARROW-HIGHWAY FONT 117.505 34.099 381
HAWTHORNE HAWT 118.369 33.923 21
LA-HABRA LAHB 117.951 33.926 82
NORTH-LONG-BEACH LGBH 118.189 33.824 7
LYNWOOD LYNN 118.210 33.929 27
PASADENA-WILSON PASA 118.127 34.134 250
PICO-RIVERA PICO 118.058 34.015 69
POMONA POMA 117.751 34.067 270
RESEDA RESE 118.533 34.199 226
RIVERSIDE-RUBIDOUX RIVR 117.417 34.000 214
SIMI-VALLEY SIMI 118.685 34.278 310
SAN-BERNARDINO SNBO 117.273 34.107 317
UPLAND-ARB UPLA 117.628 34.104 369
WHITTIER WHIT 118.025 33.924 58
WEST-LOS-ANGELES-VA-HOSPITAL WSLA 118.455 34.051 91
Figure 2.11, page 49.
Note that the x-axis in the text is reversed.
goptions reset = all;
symbol1 value=none pointlabel=("#site") color=black;
axis1 order =(117 to 119 by .4) label=(a=0 'Longitude') minor=none;
axis2 order =(33.8 to 34.3 by .1) label = (a=90 'Lattitude') minor=none ;
proc gplot data = ozone;
plot lat*long / haxis = axis1 vaxis = axis2 nolegend;
run; quit;
Figure 2.12, page 50.
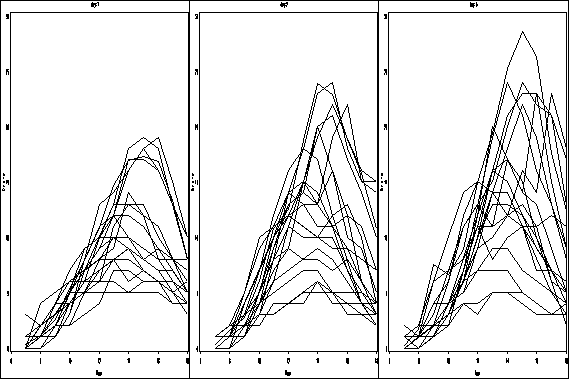
proc sort data = newozone1; by day site; run; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 20; axis1 order =(6 to 18 by 2) label=(a=0 'Hour') minor=none; axis2 order =(0 to 30 by 5) label = (a=90 'Ozone') minor=none ; proc gplot data = newozone1; by day; plot ozone*hour=site / haxis = axis1 vaxis = axis2 nolegend name = 'f2_12_'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=h3 nofs; treplay 1:f2_12_ 2:f2_12_1 3:f2_12_2; run; quit;
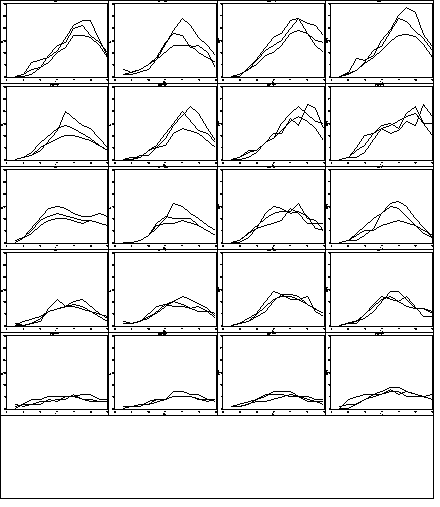
Figure 2.13, page 51.
Note that a 5×4 graph was made instead of a 4×5 graph as in the text.
proc greplay igout = work.gseg nofs;
delete _all_;
run;
data ozone1;
set newozone1;
run;
proc sql;
create table ozone2 as
select site,
max(ozone) as max_ozone
from newozone1
group by site
order by max_ozone desc, site desc;
quit;
data ozone2;
set ozone2;
order = _N_;
run;
data fmt_dataset;
retain fmtname "site";
set ozone2 ;
start = order;
label = site;
run;
proc format cntlin = fmt_dataset fmtlib;
select site;
run;
proc sql;
create table ozone3 as
select *
from ozone2, ozone1
where ozone2.site=ozone1.site
order by ozone2.order, ozone2.max_ozone, ozone2.site, ozone1.day, ozone1.hour;
quit;
goptions reset = all;
symbol1 value=none color=black interpol = join repeat = 20;
axis1 order =(6 to 18 by 2) label=(a=0 'Hour') minor=none;
axis2 order =(0 to 30 by 5) label = (a=90 'Ozone') minor=none ;
proc gplot data = ozone3;
by order;
format order site.;
plot ozone*hour=day / haxis = axis1 vaxis = axis2 nolegend name = 'f2_13_';
run; quit;
proc greplay igout = work.gseg tc=sashelp.templt template=h4 nofs ;
treplay 4:f2_13_ 3:f2_13_1 2:f2_13_2 1:f2_13_3 ;
treplay 4:f2_13_4 3:f2_13_5 2:f2_13_6 1:f2_13_7;
treplay 4:f2_13_8 3:f2_13_9 2:f2_13_10 1:f2_13_11;
treplay 4:f2_13_12 3:f2_13_13 2:f2_13_14 1:f2_13_15;
treplay 4:f2_13_16 3:f2_13_17 2:f2_13_18 1:f2_13_19;
run; quit;
proc greplay igout = work.gseg tc=sashelp.templt template = v3 nofs;
treplay 1:template 2: templat1 3:templat2;
run; quit;
proc greplay igout = work.gseg tc=sashelp.templt template = v3 nofs;
treplay 1:templat3 2:templat4;
run; quit;
proc greplay igout = work.gseg tc=sashelp.templt template = v2 nofs;
treplay 1:templat5 2:templat6;
run; quit;
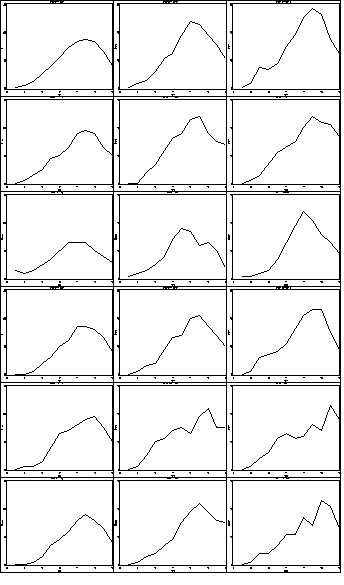
Figure 2.14, page 52.
proc greplay igout = work.gseg nofs; delete _all_; run; quit; goptions reset = all; symbol1 value=none color=black interpol = join repeat = 20; axis1 order =(6 to 18 by 2) label=(a=0 'Hour') minor=none; axis2 order =(0 to 30 by 10) label = (a=90 'Ozone') minor=none ; proc gplot data = ozone3; by order day; where order le 6; format order site.; plot ozone*hour=day / haxis = axis1 vaxis = axis2 nolegend name = 'f2_14_'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template=h3 nofs ; treplay 1:f2_14_ 2:f2_14_1 3:f2_14_2; treplay 1:f2_14_3 2:f2_14_4 3:f2_14_5; treplay 1:f2_14_6 2:f2_14_7 3:f2_14_8; treplay 1:f2_14_9 2:f2_14_10 3:f2_14_11; treplay 1:f2_14_12 2:f2_14_13 3:f2_14_14; treplay 1:f2_14_15 2:f2_14_16 3:f2_14_17; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template = v3 nofs; treplay 1:template 2: templat1 3:templat2; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template = v3 nofs; treplay 1:templat3 2:templat4 3:templat5; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template = v2 nofs; treplay 1:templat6 2:templat7; run; quit;
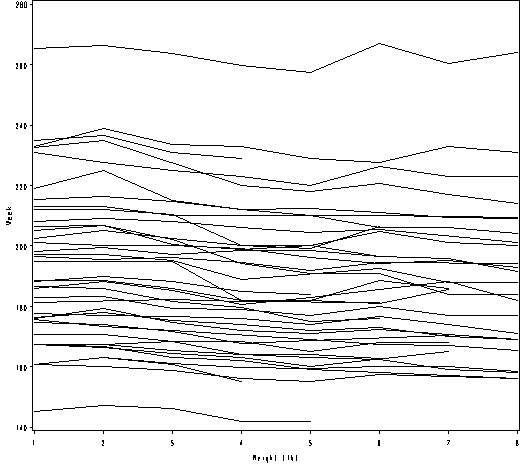
Figure 2.15, page 53.
goptions reset = all; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(1 to 8 by 1) label=(a=0 'Weight (lb)') minor=none; axis2 order =(140 to 280 by 20) label = (a=90 'Week') minor=none ; proc gplot data = weight1; plot weight*week=id / haxis = axis1 vaxis = axis2 nolegend; run; quit;
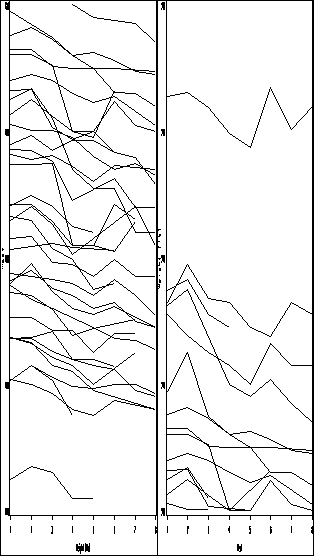
Figure 2.16, page 54.
goptions reset = all ; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(1 to 8 by 1) label=(a=0 'Weight (lb)') minor=none; axis2 order =(140 to 220 by 20) label = (a=90 'Week') minor=none ; proc gplot data = weight1; where weight le 220; plot weight*week=id / haxis = axis1 vaxis = axis2 nolegend name = 'f2_16l'; run; quit; goptions reset = all ; symbol1 value=none color=black interpol = join repeat = 38; axis1 order =(1 to 8 by 1) label=(a=0 'Week') minor=none; axis2 order =(200 to 280 by 20) label = (a=90 'Weight (lb)') minor=none ; proc gplot data = weight1; where weight ge 200; plot weight*week=id / haxis = axis1 vaxis = axis2 nolegend name = 'f2_16r'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template = h2 nofs; treplay 1:f2_16l 2:f2_16r; run; quit;
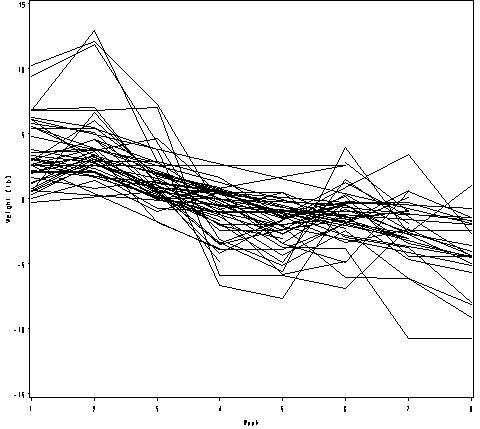
Figure 2.17, page 56.
proc sql;
create table resid_weight as
select *,
weight - mean(weight) as resid
from weight1
group by id;
quit;
goptions reset = all ;
symbol1 value=none color=black interpol = join repeat = 38;
axis1 order =(1 to 8 by 1) label=(a=0 'Week') minor=none;
axis2 order =(-15 to 15 by 5) label = (a=90 'Weight (lb)') minor=none ;
proc gplot data = resid_weight;
plot resid*week=id / haxis = axis1 vaxis = axis2 nolegend ;
run; quit;
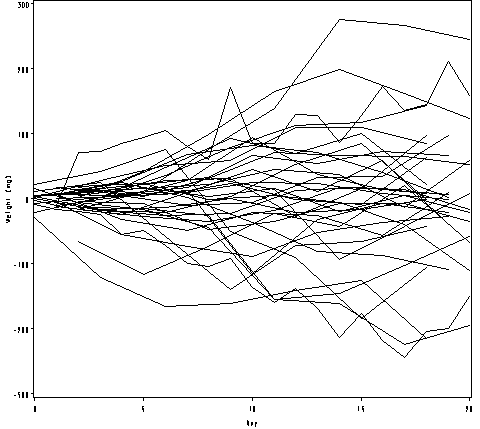
Figure 2.18, page 57.
proc sql;
create table resid_bigmice as
select *,
weight - mean(weight) as tresid
from bigmice
group by day;
quit;
goptions reset = all ;
symbol1 value=none color=black interpol = join repeat = 38;
axis1 order =(0 to 20 by 5) label=(a=0 'Day') minor=none;
axis2 order =(-300 to 300 by 100) label = (a=90 'Weight (mg)') minor=none ;
proc gplot data = resid_bigmice;
plot tresid*day=id / haxis = axis1 vaxis = axis2 nolegend ;
run; quit;
Table 2.2, page 58.
proc sort data = pain;
by id trial;
run;
proc transpose data = pain out = wide_pain prefix = l2pain ;
by id;
id trial;
var l2paintol;
run;
proc corr data = wide_pain noprob nosimple;
var l2pain1-l2pain4;
run;
The CORR Procedure
4 Variables: l2pain1 l2pain2 l2pain3 l2pain4
Pearson Correlation Coefficients
Number of Observations
l2pain1 l2pain2 l2pain3 l2pain4
l2pain1 1.00000 0.72601 0.83634 0.60431
63 62 58 60
l2pain2 0.72601 1.00000 0.71765 0.66188
62 63 58 61
l2pain3 0.83634 0.71765 1.00000 0.76424
58 58 58 58
l2pain4 0.60431 0.66188 0.76424 1.00000
60 61 58 61
Table 2.3, page 59.
proc sort data = smallmice;
by id day;
run;
proc transpose data = smallmice out = wide_smallmice prefix = weight ;
by id;
id day;
var weight;
run;
proc corr data = wide_smallmice noprob nosimple;
var weight2--weight20;
run;
The CORR Procedure
7 Variables: weight2 weight5 weight8 weight11 weight14 weight17 weight20
Pearson Correlation Coefficients, N = 14
weight2 weight5 weight8 weight11 weight14 weight17 weight20
weight2 1.00000 0.92362 0.56648 0.36127 0.23102 0.23132 0.38449
weight5 0.92362 1.00000 0.77280 0.53562 0.44996 0.40932 0.54734
weight8 0.56648 0.77280 1.00000 0.86387 0.80079 0.75786 0.80914
weight11 0.36127 0.53562 0.86387 1.00000 0.92785 0.91622 0.86744
weight14 0.23102 0.44996 0.80079 0.92785 1.00000 0.95541 0.88877
Table 2.4, page 60.
proc sort data = newozone1; by site day hour; run; proc transpose data = newozone1 out = wide_ozone prefix = ozone ; by site day; id hour; var ozone; run; proc corr data = wide_ozone noprob nosimple; var ozone7 ozone8--ozone18; run; [...output omitted...] proc corr data = wide_ozone noprob nosimple cov; var ozone7 ozone8--ozone18; run; [...output omitted...]
Figure 2.19, page 62.
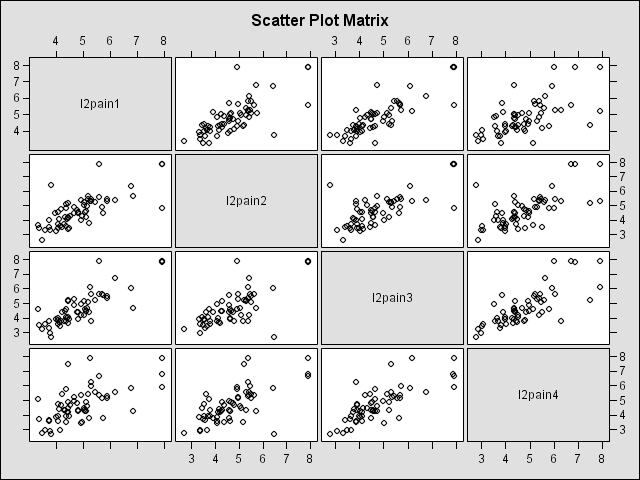
ods html; ods graphics on; proc corr data = wide_pain noprint plots = matrix; var l2pain1-l2pain4; run; ods graphics off; ods html close;
Figure 2.20, page 64.
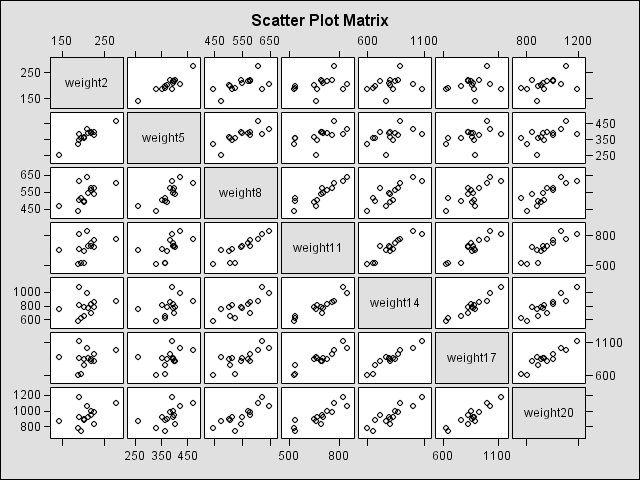
ods html; ods graphics on; proc corr data = wide_smallmice noprint plots = matrix (nmaxvar = 0); var weight2--weight20; run; ods graphics off; ods html close;
Figure 2.23, page 68.
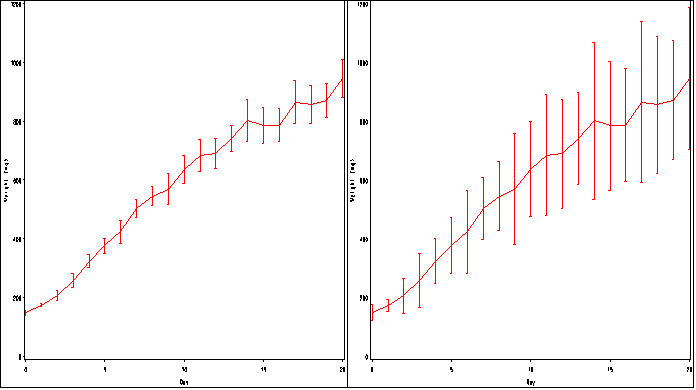
proc greplay igout = work.gseg nofs; delete _all_; run; quit; goptions reset = all ; axis1 order =(0 to 20 by 5) label=(a=0 'Day') minor=none; axis2 order =(0 to 1200 by 200) label = (a=90 'Weight (mg)') minor=none ; symbol1 interpol=std2mjt v=none color=red r=1; proc gplot data=bigmice; plot weight*day / haxis = axis1 vaxis=axis2 name = 'f2_23a'; run; quit; goptions reset = all ; axis1 order =(0 to 20 by 5) label=(a=0 'Day') minor=none; axis2 order =(0 to 1200 by 200) label = (a=90 'Weight (mg)') minor=none ; symbol1 interpol=std2jt v=none color=red r=1; proc gplot data=bigmice; plot weight*day / haxis = axis1 vaxis=axis2 name = 'f2_23b'; run; quit; proc greplay igout = work.gseg tc=sashelp.templt template = h2 nofs; treplay 1:f2_23a 2:f2_23b; run; quit;
Figure 2.24, page 69.
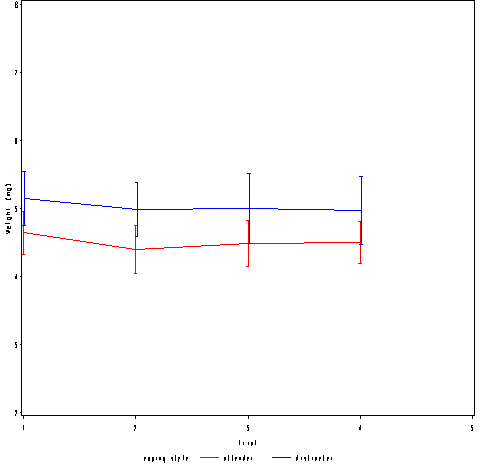
Plot A only.
data pain_offset; set pain; if cs = "distracter" then trial = trial + .01; run; goptions reset = all ; axis1 label=(a=0 'Trial') minor=none; axis2 label = (a=90 'Weight (mg)') minor=none ; symbol1 interpol=std2mjt v=none color=red r=1; symbol2 interpol=std2mjt v=none color=blue r=1; proc gplot data=pain_offset; plot l2paintol*trial = cs / haxis = axis1 vaxis=axis2 ; run; quit;
Table 2.7, page 70.
proc freq data = cognitive; where ravens ne . ; table round; run;
The FREQ Procedure
Cumulative Cumulative
ROUND Frequency Percent Frequency Percent
1 543 21.44 543 21.44
2 510 20.13 1053 41.57
3 509 20.09 1562 61.67
4 497 19.62 2059 81.29
5 474 18.71 2533 100.00
Table 2.8, page 70.
proc sql;
create table cog as
select id,
sum((ravens ne .)) as observed_raven
from cognitive
group by id;
quit;
proc freq data = cog;
table observed_raven;
run;
The FREQ Procedure
Cumulative Cumulative
observed_raven Frequency Percent Frequency Percent
1 8 1.48 8 1.48
2 21 3.89 29 5.37
3 16 2.96 45 8.33
4 40 7.41 85 15.74
5 455 84.26 540 100.00
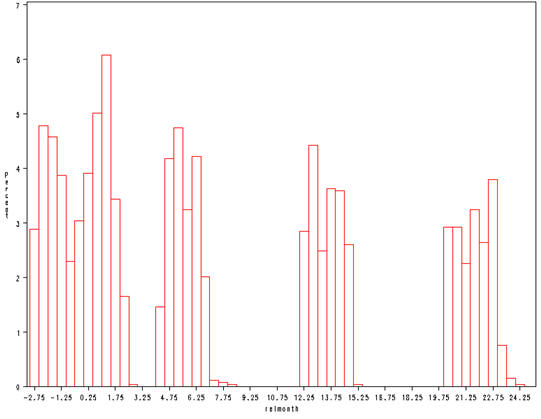
Figure 2.25, page 71.
proc univariate data = cognitive noprint; histogram relmonth / midpoints=-2.75 to 24.75 by .5; run;
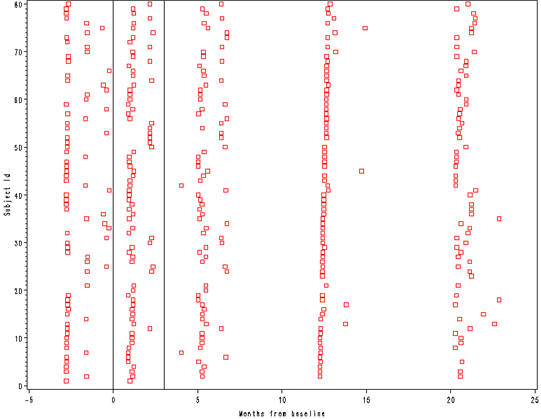
Figure 2.26, page 72.
goptions reset = all ; axis1 label=(a=0 'Months from baseline') order = (-5 to 25 by 5) minor=none; axis2 label = (a=90 'Subject Id'); symbol1 value=square color=red; proc gplot data=cognitive; where id le 80; plot id*relmonth / haxis = axis1 vaxis=axis2 href = 0 3; run; quit;