In this chapter we create and use the variables GndC_verb which is equal to iq_verb centered around the grand mean; GrpMC_verb which contains the group means of GndC_verb, so it contains the group means of iq_verb centered around the grand mean.
Table 4.1, p. 47.
The random intercept only model.
proc mixed data=schools covtest noclprint noitprint method=ml;
class schoolnr;
model langpost = / solution;
random intercept / subject=schoolnr;
run;
<output omitted>
Covariance Parameter Estimates
Standard Z
Cov Parm Subject Estimate Error Value Pr Z
Intercept schoolNR 19.4126 3.0962 6.27 <.0001
Residual 64.5704 1.9729 32.73 <.0001
Fit Statistics
-2 Log Likelihood 16253.2
AIC (smaller is better) 16259.2
AICC (smaller is better) 16259.2
BIC (smaller is better) 16267.8
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 40.3642 0.4262 130 94.70 <.0001
Creating the variable GndC_verb which is equal to iq_verb centered around the grand mean.
proc sql; create table schools1 as select *, iq_verb - mean(iq_verb) as GndC_verb from schools; quit;
Table 4.2, p. 49.
Random intercept model with effect for IQ centered around the grand mean.
proc mixed data=schools1 covtest noclprint noitprint;
class schoolnr;
model langpost = GndC_verb / solution;
random intercept / subject=schoolnr;
run;
<output omitted>
Covariance Parameter Estimates
Standard Z
Cov Parm Subject Estimate Error Value Pr Z
Intercept schoolNR 9.6015 1.5927 6.03 <.0001
Residual 42.2446 1.2893 32.77 <.0001
Fit Statistics
-2 Res Log Likelihood 15255.8
AIC (smaller is better) 15259.8
AICC (smaller is better) 15259.8
BIC (smaller is better) 15265.5
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 40.6082 0.3082 130 131.77 <.0001
GndC_verb 2.4876 0.07008 2155 35.50 <.0001
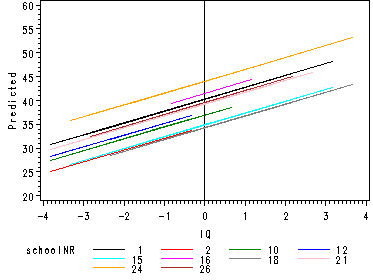
Fig. 4.2, p. 49.
Randomly chosen regression lines according to the random intercept model of table 4.4.
proc mixed data=schools2 covtest noitprint noclprint method=ml;
class schoolnr;
model langpost = GndC_verb / outp=p ;
random intercept / subject=schoolnr type=un;
run;
goptions reset=all;
symbol i=j r=24;
axis1 order=(-4 to 4 by 1) label=('IQ');
axis2 order=(20 to 60 by 5) label=(a=90 'Predicted');
proc gplot data=p;
where schoolnr < 27;
plot pred*GndC_verb = schoolnr / vaxis=axis2 haxis=axis1 href=0 ;
run;
quit;
Table 4.3, p. 51.
Ordinary least squares regression of the model in table 4.2.
proc reg data=schools1;
model langpost = GndC_verb;
run;
quit;
Analysis of Variance
Sum of Mean
Source DF Squares Square F Value Pr > F
Model 1 68916 68916 1352.84 <.0001
Error 2285 116402 50.94159
Corrected Total 2286 185317
Root MSE 7.13734 R-Square 0.3719
Dependent Mean 40.93485 Adj R-Sq 0.3716
Coeff Var 17.43585
Parameter Estimates
Parameter Standard
Variable DF Estimate Error t Value Pr > |t|
Intercept 1 40.93485 0.14925 274.28 <.0001
GndC_verb 1 2.65390 0.07215 36.78 <.0001
Table 4.4, p. 55.
Random intercept model with different within and between group regressions.
Note: The “IQ here is the raw variable” means that we are using the grand mean centered variable GndC_verb and not iq_verb. Furthermore, the group mean variable for IQ is actually the group mean variable of GndC_verb which we here have called GrpMC_verb.
*Group mean centering of GndC_verb. ;
proc sql;
create table schools2 as
select *, mean(GndC_verb) as GrpMC_verb
from schools1
group by schoolnr;
quit;
proc mixed data=schools2 covtest noitprint noclprint method=ml;
class schoolnr;
model langpost = GndC_verb GrpMC_verb / solution;
random intercept / subject=schoolnr;
run;
<output omitted>
Covariance Parameter Estimates
Standard Z
Cov Parm Subject Estimate Error Value Pr Z
Intercept schoolNR 7.7275 1.3093 5.90 <.0001
Residual 42.1519 1.2842 32.82 <.0001
Fit Statistics
-2 Log Likelihood 15227.5
AIC (smaller is better) 15237.5
AICC (smaller is better) 15237.6
BIC (smaller is better) 15251.9
Solution for Fixed Effects
Standard
Effect Estimate Error DF t Value Pr > |t|
Intercept 40.7423 0.2844 129 143.26 <.0001
GndC_verb 2.4148 0.07166 2155 33.70 <.0001
GrpMC_verb 1.5885 0.3127 2155 5.08 <.0001
The remaining tables and figures have been skipped.