The aim of this seminar is to help you increase your skills in analyzing repeated measures data using SAS. The seminar will describe conventional ways to analyze repeated measures using SAS PROC GLM and describe the assumptions and limitations of such conventional methods. Then we will explore the use of SAS PROC MIXED for repeated measures analyses.
SAS Program for seminar.
There are a number of situations that can arise when the analysis includes between groups effects as well as within subject effects. We start by showing 4 example analyses using measurements of depression over 3 time points broken down by 2 treatment groups. In the first example we see that the two groups differ in depression but neither group changes over time. In the second example the two groups grow in depression but at the same rate over time. In the third example, the two groups start off being quite different in depression but end up being rather close in depression. The fourth example shows the groups starting off at the same level of depression, and one group group increases over time whereas the other group decreases over time.
Note that in the interest of making learning the concepts easier we have taken the liberty of using only a very small portion of the voluminous output that SAS provides and we have inserted the graphs as needed to facilitate understanding the concepts. The rather long code needed to actually create the graphs in SAS has been included at the very end after all the output and explanation.
Demo Analysis #1
The between groups test indicates that there the variable group is significant, consequently in the graph we see that the lines for the two groups are rather far apart. The within subject test indicate that there is not a significant time effect, in other words, the groups do not change in depression over time. In the graph we see that the groups have lines that are flat, i.e. the slopes of the lines are approximately equal to zero. Also, since the lines are parallel, we are not surprised that the interaction between time and group is not significant.
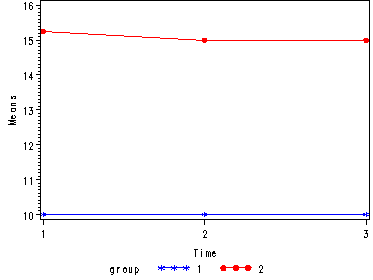
<Abbreviated output from proc glm> Tests of Hypotheses for Between Subjects Effects Source DF Type III SS Mean Square F Value Pr > F GROUP 1 155.0416667 155.0416667 3721.00 <.0001 Error 6 0.2500000 0.0416667 Univariate Tests of Hypotheses for Within Subject Effects Source DF Type III SS Mean Square F Value Pr > F time 2 0.08333333 0.04166667 1.00 0.3966 time*GROUP 2 0.08333333 0.04166667 1.00 0.3966 Error(time) 12 0.50000000 0.04166667
Demo Analysis #2
The between groups test indicates that there the variable group is not significant, consequently in the graph we see that the lines for the two groups are rather close together. The within subject test indicate that there is a significant time effect, in other words, the groups do change in depression over time. In the graph we see that the groups have lines that increase over time. Again, the lines are parallel consistent with the finding that the interaction is not significant.
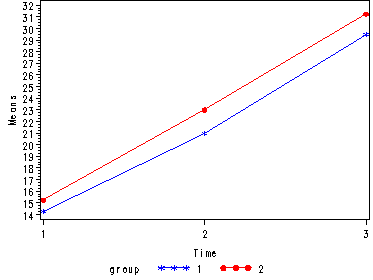
<Abbreviated output from proc glm>
Tests of Hypotheses for Between Subjects Effects Source DF Type III SS Mean Square F Value Pr > F GROUP 1 15.0416667 15.0416667 0.84 0.3957 Error 6 107.9166667 17.9861111 Univariate Tests of Hypotheses for Within Subject Effects Source DF Type III SS Mean Square F Value Pr > F time 2 978.2500000 489.1250000 53.68 <.0001 time*GROUP 2 1.0833333 0.5416667 0.06 0.9426 Error(time) 12 109.3333333 9.1111111
Demo Analysis #3
The between groups test indicates that there the variable group is significant, consequently in the graph we see that the lines for the two groups are rather far apart. The within subject test indicate that there is a significant time effect, in other words, the groups do change over time, both groups are getting less depressed over time. Moreover, the interaction of time and group is significant which means that the groups are changing over time but are changing in different ways, which means that in the graph the lines will not be parallel. In the graph we see that the groups have non-parallel lines that decrease over time and are getting progressively closer together over time.
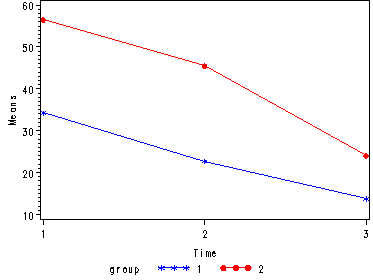
Tests of Hypotheses for Between Subjects Effects Source DF Type III SS Mean Square F Value Pr > F GROUP 1 2035.041667 2035.041667 343.15 <.0001 Error 6 35.583333 5.930556 Univariate Tests of Hypotheses for Within Subject Effects Source DF Type III SS Mean Square F Value Pr > F time 2 2830.333333 1415.166667 553.76 <.0001 time*GROUP 2 200.333333 100.166667 39.20 <.0001 Error(time) 12 30.666667 2.555556
Demo Analysis #4
The within subject test indicate that the interaction of time and group is significant. The main effect of time is not significant. However, the significant interaction indicates that the groups are changing over time and they are changing in different ways, in other words, in the graph the lines of the groups will not be parallel. The between groups test indicates that there the variable group is significant. In the graph for this particular case we see that one group is increasing in depression over time and the other group is decreasing in depression over time.
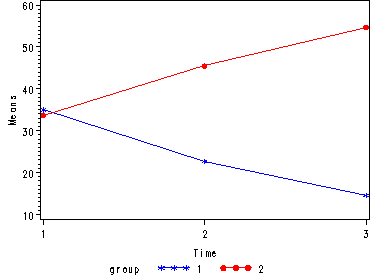
Tests of Hypotheses for Between Subjects Effects Source DF Type III SS Mean Square F Value Pr > F GROUP 1 2542.041667 2542.041667 628.96 <.0001 Error 6 24.250000 4.041667 Univariate Tests of Hypotheses for Within Subject Effects Source DF Type III SS Mean Square F Value Pr > F time 2 1.000000 0.500000 0.08 0.9246 time*GROUP 2 1736.333333 868.166667 137.08 <.0001 Error(time) 12 76.000000 6.333333
Creating Graphs of the Means for Demo Analysis #4
The SAS code for creating the graph for demo=4.
/* We use the out option in the lsmeans statement to create the data set means. */ proc glm data=demo4; class group; model time1 time2 time3 = group; repeated time 3 ; lsmeans group / out=means; run; quit; /*We want to look at the means to make sure we created the correct dataset.*/ proc print data=means; run; /* For a better understanding of all the gplot options used here please visit our webpage on using proc gplot.*/ goptions reset=all; symbol1 c=blue v=star h=.8 i=j; symbol2 c=red v=dot h=.8 i=j; axis1 label=(a=90 'Means'); axis2 label=('Time') value=('1' '2' '3'); proc gplot data=means; plot lsmean*_name_=group/ vaxis=axis1 haxis=axis2; run; quit;
Exercise data examples
The data consists of people who were randomly assigned to two different diets: low-fat and not low-fat and three different types of exercise: at rest, walking leisurely and running. Their pulse rate was measured at three different time points during their assigned exercise: at 1 minute, 15 minutes and 30 minutes.
data exercise; input id exertype diet time1 time2 time3; cards; 1 1 1 85 85 88 2 1 1 90 92 93 3 1 1 97 97 94 4 1 1 80 82 83 5 1 1 91 92 91 6 1 2 83 83 84 7 1 2 87 88 90 8 1 2 92 94 95 9 1 2 97 99 96 10 1 2 100 97 100 11 2 1 86 86 84 12 2 1 93 103 104 13 2 1 90 92 93 14 2 1 95 96 100 15 2 1 89 96 95 16 2 2 84 86 89 17 2 2 103 109 90 18 2 2 92 96 101 19 2 2 97 98 100 20 2 2 102 104 103 21 3 1 93 98 110 22 3 1 98 104 112 23 3 1 98 105 99 24 3 1 87 132 120 25 3 1 94 110 116 26 3 2 95 126 143 27 3 2 100 126 140 28 3 2 103 124 140 29 3 2 94 135 130 30 3 2 99 111 150 ; run;
Exercise example, model 1 (time and diet)
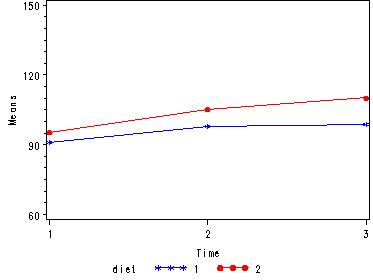
Let us first consider the model including diet as the group variable. The graph below suggests that the pulse rate is growing over time. The pulse rates may vary for the 2 diets and it is possible that the pulse rate is growing faster for the “red” diet than the “blue” diet.
proc glm data=exercise; class diet; model time1 time2 time3 = diet; repeated time 3 / printe; run; quit;
Looking at the results from the manova test the effect of time is significant but the interaction of time and diet is not significant. The between subject test of the effect of diet is also not significant. Consequently, in the graph we have lines that are not flat, in fact, they are actually increasing over time, which was expected since the effect of time was significant. Furthermore, the lines are approximately parallel which was anticipated since the interaction was not significant.
Sphericity Tests
Mauchly's
Variables DF Criterion Chi-Square Pr > ChiSq
Transformed Variates 2 0.4531199 21.373158 <.0001
Orthogonal Components 2 0.673336 10.678793 0.0048
Manova Test Criteria and Exact F Statistics for the Hypothesis of no time Effect
H = Type III SSCP Matrix for time
E = Error SSCP Matrix
S=1 M=0 N=12.5
Statistic Value F Value Num DF Den DF Pr > F
Wilks' Lambda 0.64349965 7.48 2 27 0.0026
Pillai's Trace 0.35650035 7.48 2 27 0.0026
Hotelling-Lawley Trace 0.55400240 7.48 2 27 0.0026
Roy's Greatest Root 0.55400240 7.48 2 27 0.0026
Manova Test Criteria and Exact F Statistics
for the Hypothesis of no time*DIET Effect
H = Type III SSCP Matrix for time*DIET
E = Error SSCP Matrix
S=1 M=0 N=12.5
Statistic Value F Value Num DF Den DF Pr > F
Wilks' Lambda 0.94402156 0.80 2 27 0.4595
Pillai's Trace 0.05597844 0.80 2 27 0.4595
Hotelling-Lawley Trace 0.05929784 0.80 2 27 0.4595
Roy's Greatest Root 0.05929784 0.80 2 27 0.4595
Tests of Hypotheses for Between Subjects Effects
Source DF Type III SS Mean Square F Value Pr > F
DIET 1 1261.87778 1261.87778 3.15 0.0869
Error 28 11227.02222 400.96508
Repeated Measures Analysis of Variance
Univariate Tests of Hypotheses for Within Subject Effects
Source DF Type III SS Mean Square F Value Pr > F
time 2 2066.600000 1033.300000 11.81 <.0001
time*diet 2 192.822222 96.411111 1.10 0.3394
Error(time) 56 4900.577778 87.510317
Adj Pr > F
Source G - G H - F
time 0.0003 0.0002
time*diet 0.3264 0.3303
Error(time)
Greenhouse-Geisser Epsilon 0.7538
Huynh-Feldt Epsilon 0.8158
Exercise example, model 2 (time and exercise type)
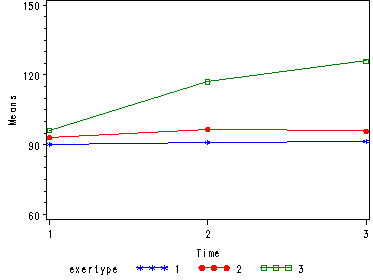
Next, let us consider the model including exertype as the group variable.
proc glm data=exercise; class exertype; model time1 time2 time3 = exertype; repeated time 3 ; run; quit;
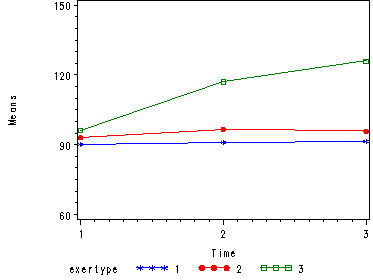
The interaction of time and exertype is significant as is the effect of time. The between subject test of the effect of exertype is also significant. Consequently, in the graph we have lines that are not parallel which we expected since the interaction was significant. Furthermore, we see that some of the lines that are rather far apart and at least one line is not horizontal which was anticipated since exertype and time were both significant. The output for this analysis is omitted.
Here is the code for the graph.
proc glm data=exercise;
class exertype;
model time1 time2 time3 = exertype;
repeated time 3 ;
lsmeans exertype / out=means;
run;
quit;
proc print data=means;
run;
goptions reset=all;
symbol1 c=blue v=star h=.8 i=j;
symbol2 c=red v=dot h=.8 i=j;
symbol3 c=green v=square h=.8 i=j;
axis1 order=(60 to 150 by 30) label=(a=90 'Means');
axis2 label=('Time') value=('1' '2' '3');
proc gplot data=means;
plot lsmean*_name_=exertype / vaxis=axis1 haxis=axis2;
run;
quit;
Further Issues
Missing Data
- Compare GLM and Mixed on Missing Data
Variance-Covariance Structures
- Discuss “univariate” vs. “multivariate” tests.
- Discuss “sphericity” and test of sphericity.
Independence
As though analyzed using between subjects analysis.
s2 0 s2 0 0 s2
Compound Symmetry
The univariate tests assumes that the variance-covariance structure has compound symmetry. There is a single Variance (represented by s2) for all 3 of the time points and there is a single covariance (represented by s1) for each of the pairs of trials. This is illustrated below.
s2 s1 s2 s1 s1 s2
Unstructured
The manova tests assumes that each variance and covariance is unique, see below, referred to as an unstructured
covariance matrix. Each trial has its own variance (e.g. s12 is the variance of trial 1) and each pair of trials has its own covariance (e.g. s21 is the covariance of trial 1 and trial2).
s12 s21 s22 s31 s32 s32
We can use the sphericity test to indicate which is most appropriate: the manova or the univariate test. The null hypothesis test of the test of sphericity is: the variance-covariance structure has the Huynh-Feldt structure, so called Type H structure. If the sphericity test is not significant then we can not reject that null hypothesis that the variance-covariance structure has Type H structure. A Type H structure is a variance-covariance structure more general than compound symmetry structure that allows the use of the univariate tests. If, however, the sphericity test is significant then we reject that the variance-covariance structure has a Type H structure and it is most appropriate to use the results from the manova test or alternatively use the corrections for the univariate test. It is very important, however, to note that the sphericity test is overly sensitive. It is very likely to reject compound symmetry when the data only slightly deviates from compound symmetry, so in actuality this test could be very deceiving and may be best ignored.
Autoregressive
Another common covariance structure which is frequently observed in repeated measures data is an autoregressive structure, which recognizes that observations which are more proximate are more correlated than measures that are more distant.
s2 sr s2 sr2 sr s2
Autoregressive Heterogeneous Variances
If the variances change over time, then the covariance would look like this.
s12 sr s22 sr2 sr s32
However, we cannot use this kind of covariance structure in a traditional repeated measures analysis, but we can use SAS PROC MIXED for such an analysis.
Let’s look at the correlations, variances and covariances for the exercise data.
proc corr data=exercise cov;
var time1 time2 time3;
run;
Covariance Matrix, DF = 29
time1 time2 time3
time1 37.8436782 48.7885057 60.2850575
time2 48.7885057 212.1195402 233.7609195
time3 60.2850575 233.7609195 356.3229885
Pearson Correlation Coefficients, N = 30
time1 time2 time3
time1 1.00000 0.54454 0.51915
time2 0.54454 1.00000 0.85028
time3 0.51915 0.85028 1.00000
SAS Exercise example, model 2 using Proc Mixed
Even though we are very impressed with our results so far, we are not completely convinced that the variance-covariance structure really has compound symmetry. In order to compare models with different variance-covariance structures we have to use proc mixed and try the different structures that we think our data might have. However, in order to use proc mixed we must reshape our data from its wide form to a long form.
proc transpose data=exercise out=long; by id diet exertype; run; data long; set long (rename=(col1=pulse) ); time = substr(_NAME_, 5, 1 )+0; drop _name_; run; proc print data=long (obs=20); var id diet exertype time pulse; run; Obs id DIET EXERTYPE time pulse 1 1 1 1 1 85 2 1 1 1 2 85 3 1 1 1 3 88 4 2 1 1 1 90 5 2 1 1 2 92 6 2 1 1 3 93 7 3 1 1 1 97 8 3 1 1 2 97 9 3 1 1 3 94 10 4 1 1 1 80 11 4 1 1 2 82 12 4 1 1 3 83 13 5 1 1 1 91 14 5 1 1 2 92 15 5 1 1 3 91 16 6 2 1 1 83 17 6 2 1 2 83 18 6 2 1 3 84 19 7 2 1 1 87 20 7 2 1 2 88
Compound Symmetry
The first model we will look at is one using compound symmetry for the variance-covariance structure. This model should confirm the results of the univariate tests that we obtained through proc glm and we will be able to obtain fit statistics that we will use for comparisons with our models that assume other variance-covariance structures.
proc mixed data=long;
class exertype time;
model pulse = exertype time exertype*time;
repeated time / subject=id type=cs;
run;
Fit Statistics
-2 Res Log Likelihood 590.8
AIC (smaller is better) 594.8
AICC (smaller is better) 595.0
BIC (smaller is better) 597.6
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
1 15.36 <.0001
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
exertype 2 27 27.00 <.0001
time 2 54 23.54 <.0001
exertype*time 4 54 15.51 <.0001
Unstructured
We now try an unstructured covariance matrix.
proc mixed data=long;
class exertype time;
model pulse = exertype time exertype*time;
repeated time / subject=id type=un;
run;
Covariance Parameter Estimates
Cov Parm Subject Estimate
UN(1,1) id 34.2000
UN(2,1) id 23.6852
UN(2,2) id 87.1926
UN(3,1) id 26.7889
UN(3,2) id 59.8148
UN(3,3) id 120.57
Fit Statistics
-2 Res Log Likelihood 577.7
AIC (smaller is better) 589.7
AICC (smaller is better) 590.9
BIC (smaller is better) 598.1
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
5 28.46 <.0001
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
exertype 2 27 27.00 <.0001
time 2 27 22.32 <.0001
exertype*time 4 27 14.39 <.0001
Autoregressive
From previous studies we suspect that our data might actually have an auto-regressive variance-covariance structure so this is the model we will look at next. The auto-regressive variance-covariance structure does fit our data slightly better than the compound symmetry does (AIC of 594.1 vs. 594.8).
proc mixed data=long;
class exertype time;
model pulse = exertype time exertype*time;
repeated time / subject=id type=ar(1);
run;
-2 Res Log Likelihood 590.1
AIC (smaller is better) 594.1
AICC (smaller is better) 594.3
BIC (smaller is better) 596.9
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
1 16.08 <.0001
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
exertype 2 27 28.39 <.0001
time 2 54 18.20 <.0001
exertype*time 4 54 11.73 <.0001
Autoregressive with heterogeneous variances
Now we suspect that what is actually going on is that the we have auto-regressive covariances and heterogeneous variances. The fit statistics indicate that our suspicions were correct (see table in Model Comparisons section) and that the model with heterogeneous variances fits the data better than the model with autoregressive covariance and homogeneous variances (AIC 587.8 versus 594.1). Our suspicions arose when we were looking at the raw covariance structure obtained from the proc corr. When looking at the output we see that the variances (the numbers along the diagonal) are clearly unequal indicating heterogeneous variances.
proc mixed data=long;
class exertype time;
model pulse = exertype time exertype*time;
repeated time / subject=id type=arh(1);
run;
Covariance Parameter Estimates
Cov
Parm Subject Estimate
Var(1) id 35.7683
Var(2) id 87.1927
Var(3) id 115.50
ARH(1) id 0.5101
Fit Statistics
-2 Res Log Likelihood 579.8
AIC (smaller is better) 587.8
AICC (smaller is better) 588.3
BIC (smaller is better) 593.4
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
3 26.42 <.0001
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
exertype 2 27 28.96 <.0001
time 2 54 21.92 <.0001
exertype*time 4 54 13.81 <.0001
It is very important to explore different variance-covariance structures when using proc mixed because the output contains fit statistics indicating which clearly indicate how well each model fits the data compared to other models.
Model comparison (comparing to Compound Symmetry)
| Model | AIC | -2RLL | Parms (df + 1) | Diff -2RLL (vs. CS) | Diff in df (vs. CS) | p value for Diff (from a chi square dist) |
| Compound Symmetry | 594.8 | 590.8 | 2 | |||
| Unstructured | 589.7 | 577.7 | 6 | 13.1 | 4 | .01 |
| Autoregressive | 594.1 | 590.1 | 2 | .7 | 0 | na |
| Autoregressive Heterogenous Variances | 587.8 | 579.8 | 4 | 11 | 2 | 0.004 |
The two most promising structures are Autoregressive Heterogeneous Variances and Unstructured since these two models have the smallest AIC values and the -2 Log Likelihood scores are significantly smaller than the -2 Log Likehood scores of other models.
Creating Graphs of the Means for Proc Mixed, model 2 (time and exertype)
Just as in the case of proc glm it is often very useful to look at the graph of the means in order to really understand the data. So, here is the code for creating the graphs in proc mixed that we were able to obtain when using proc glm.
/* Proc Mixed does not have an out option in the lsmeans statement. Instead we use ODS to create the data set containing all the means. */ ods output LSMeans=means1; proc mixed data=long; class exertype time; model pulse = exertype time exertype*time; repeated time / subject=id type=ar(1); lsmeans time*exertype; run; /* We print the dataset just to make sure that we have created the correct dataset. */ proc print data=means1; run; /* First we reset all the plot options to avoid any carry over from previous plotting procedures. We use a format statement in the proc gplot because the values for estimate have been assigned many decimal places that do not look very nice when used as tick marks on the y-axis. The format 8. means that we will allow there to be 8 digits for the whole number and no decimal places. This statement is included purely for cosmetic purposes and can easily be removed. To understand all the plotting options used please refer to our webpage on using proc gplot. */ goptions reset=all; symbol1 c=blue v=star h=.8 i=j; symbol2 c=red v=dot h=.8 i=j; symbol3 c=green v=square h=.8 i=j; axis1 order=(60 to 150 by 30) label=(a=90 'Means'); proc gplot data=means1; format estimate 8.; plot estimate*time=exertype / vaxis=axis1; run; quit;
Here is the graph.
Exercise example, model 3 (time, diet and exertype)–Proc Glm
Looking at models including only diet or exertype separately does not answer all our questions. We would also like to know if the people on the low-fat diet who engage in running have lower pulse rates than the people participating in the not low-fat diet who are not running. In order to address these types of questions we need to look at a model that includes the interaction of diet and exertype. After all the analysis involving the variance-covariance structures we will look at this model using both proc glm and proc mixed.
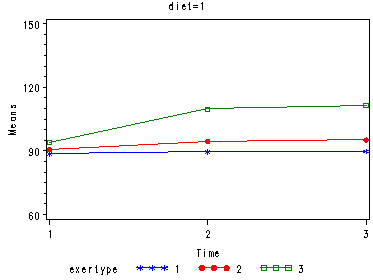
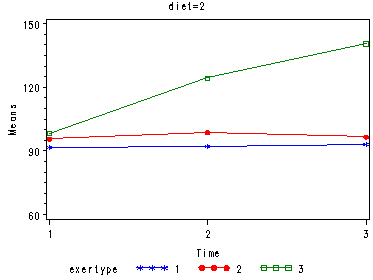
In the graph of exertype by diet we see that for the low-fat diet (diet=1) group the pulse rate for the two exercise types: at rest and walking, are very close together, indeed they are almost flat, whereas the running group has a higher pulse rate that increases over time. For the not low-fat diet (diet=2) group the same two exercise types: at rest and walking, are also very close together and almost flat. For this group, however, the pulse rate for the running group increases greatly over time and the rate of increase is much steeper than the increase of the running group in the low-fat diet group.
The within subject tests indicate that there is a three-way interaction between diet, exertype and time. In other words, the pulse rate will depend on which diet you follow, the exercise type you engage in and at what time during the exercise that you measure the pulse. The interactions of time and exertype and diet and exertype are also significant as are the main effects of diet and exertype.
proc glm data=exercise; class diet exertype; model time1 time2 time3 = diet|exertype; repeated time 3 ; run; quit;
Looking at the graphs of exertype by diet.
Tests of Hypotheses for Between Subjects Effects Source DF Type III SS Mean Square F Value Pr > F DIET 1 1261.877778 1261.877778 14.52 0.0008 EXERTYPE 2 8326.066667 4163.033333 47.92 <.0001 DIET*EXERTYPE 2 815.755556 407.877778 4.69 0.0190 Error 24 2085.200000 86.883333 Univariate Tests of Hypotheses for Within Subject Effects Source DF Type III SS Mean Square F Value Pr > F time 2 2066.600000 1033.300000 31.72 <.0001 time*DIET 2 192.822222 96.411111 2.96 0.0614 time*EXERTYPE 4 2723.333333 680.833333 20.90 <.0001 time*DIET*EXERTYPE 4 613.644444 153.411111 4.71 0.0028 Error(time) 48 1563.600000 32.575000
Creating Graphs for model 3 Using Proc Glm
proc glm data=exercise;
class diet exertype;
model time1 time2 time3 = diet|exertype;
repeated time 3;
lsmeans diet*exertype / out=means;
run;
quit;
proc print data=means;
run;
proc sort data=means out=sortdiet;
by diet;
run;
goptions reset=all;
symbol1 c=blue v=star h=.8 i=j;
symbol2 c=red v=dot h=.8 i=j;
symbol3 c=green v=square h=.8 i=j;
axis1 order=(60 to 150 by 30) label=(a=90 'Means');
axis2 label=('Time') value=('1' '2' '3');
proc gplot data=sortdiet;
by diet;
plot lsmean*_name_ = exertype / vaxis=axis1 haxis=axis2;
run;
quit;
Exercise example, model 3 (time, diet and exertype)–Proc Mixed
For the mixed model we will use the autoregressive heterogeneous variances variance-covariance structure since we previously observed that this is the structure that appears to fit the data the best (see discussion of variance-covariance structures). We do not expect to find a great change in which factors will be significant but we do expect to have a model that has a better fit than the glm model. The graphs are exactly the same as the glm model and we find that the same factors are significant. However, since the model has a better fit we can be more confident in the estimate of the standard errors and therefore we can be more confident in the tests and in the findings of significant factors. The model has a better fit than the model only including exertype and time because both the -2Log Likelihood and the AIC has decrease dramatically. The -2 Log Likelihood decreased from 579.8 for the model including only exertype and time to 505.3 for the current model; the AIC decreased from 587.8 for the model including only exertype and time to 513.3 for the current model.
The code for the mixed model proc mixed data=long; class exertype diet time; model pulse = exertype|diet|time; repeated time / subject=id type=arh(1) ; run;
Looking at the graphs of exertype by diet.
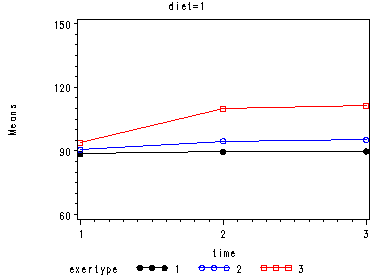
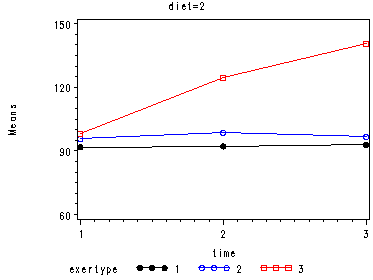
Covariance Parameter Estimates
Cov
Parm Subject Estimate
Var(1) id 33.0864
Var(2) id 73.5148
Var(3) id 45.3847
ARH(1) id 0.3610
Fit Statistics
-2 Res Log Likelihood 505.3
AIC (smaller is better) 513.3
AICC (smaller is better) 513.9
BIC (smaller is better) 518.9
Null Model Likelihood Ratio Test
DF Chi-Square Pr > ChiSq
3 10.65 0.0138
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
exertype 2 24 52.17 <.0001
diet 1 24 15.81 0.0006
exertype*diet 2 24 5.11 0.0142
time 2 48 30.82 <.0001
exertype*time 4 48 20.25 <.0001
diet*time 2 48 2.80 0.0709
exertype*diet*time 4 48 4.45 0.0039
Creating Graphs for model 3 Using Proc Mixed
/* Proc Mixed does not have an out option in the lsmeans statement. Instead we use ODS to create the data set containing all the means. */ ods output LSMeans = means; proc mixed data=long; class exertype diet time; model pulse = exertype|diet|time; repeated time / subject=id type=arh(1) ; lsmeans time*diet*exertype; run; /* We print the dataset just to make sure that we have created the correct dataset. */ proc print data=means; run; proc sort data=means; by diet; run; /* First we reset all the plot options to avoid any carry over from previous plotting procedures. We use a format statement in the proc gplot because the values for estimate have been assigned many decimal places that do not look very nice when used as tick marks on the y-axis. The format 8. means that we will allow there to be 8 digits for the whole number and no decimal places. This statement is included purely for cosmetic purposes and can easily be removed. To understand all the plotting options used please refer to our webpage on using proc gplot. */ goptions reset=all; symbol1 c=black v=dot i=j; symbol2 c=blue v=circle i=j; symbol3 c=red v=square i=j; axis1 order=(60 to 150 by 30) label=(a=90 'Means'); proc gplot data=means; by diet; format estimate 8.; plot estimate*time=exertype / vaxis=axis1; run; quit;
Contrasts and interaction contrasts for model 3
From the graphs in the above analysis we see that the runners (exertype level 3) have a pulse rate that is increases much quicker than the pulse rates of the two other groups. We would like to know if there is a statistically significant difference between the changes over time in the pulse rate of the runners versus the change over time in the pulse rate of the walkers and the “rest-ers” (the people at rest) across diet groups and across time. Furthermore, we suspect that there might be a difference in pulse rate over time and across exercise type between the two diet groups. But to make matters even more complicated we would like to test if the runners in the low fat diet group are statistically significantly different from all the other groups (i.e. the runners in the non-low fat diet, the walkers and the “rest-ers” in both diet groups). Since we are being ambitious we also want to test if the runners in the low fat diet group (diet=1) are different from the runners in the non-low fat diet group (diet=2). These contrasts are all tested using the estimate statement in proc mixed.
If we would like to look at the differences among groups at each level of another variable we have to utilize the lsmeans statement with the slice option. For example, we could test for differences among the exertype groups at each level of diet across all levels of time; or we could test for differences in groups of exertype for each time point across both levels of diet; we could also test for differences in groups of exertype for each combination of time and diet levels.
proc mixed data=long;
class diet exertype time;
model pulse = exertype|diet|time ;
repeated time / subject=id type=arh(1) ;
estimate 'exer 12 v 3' exertype -.5 -.5 1; /* across time and across diet groups */
estimate 'exer 1 v 2' exertype -1 1 0; /* across time and across diet groups */
estimate 'diet' diet -1 1; /* across time and across exercise types */
estimate 'diet 1v2 & exertype 12v3'
diet*exertype -.5 -.5 1
.5 .5 -1; /* across time only */
estimate 'runners only, diet 1 v 2' diet 1 -1
diet*exertype 0 0 1
0 0 -1; /* across time only */
lsmeans diet*exertype / slice=diet; /*testing for differences among exertype for each level of diet */
lsmeans exertype*time / slice=time; /*differences in exertype for each time point*/
lsmeans exertype*diet*time / slice=time*diet;
run;
quit;
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
exertype 2 24 52.17 <.0001
diet 1 24 15.81 0.0006
diet*exertype 2 24 5.11 0.0142
time 2 48 30.82 <.0001
exertype*time 4 48 20.25 <.0001
diet*time 2 48 2.80 0.0709
diet*exertype*time 4 48 4.45 0.0039
Estimates
Standard
Label Estimate Error DF t Value Pr > |t|
exer 12 v 3 20.0500 1.9975 24 10.04 <.0001
exer 1 v 2 4.3667 2.3066 24 1.89 0.0705
diet 7.4889 1.8833 24 3.98 0.0006
diet 1v2 & exertype 12v3 -12.7667 3.9951 24 -3.20 0.0039
runners only, diet 1 v 2 -16.0000 3.2620 24 -4.91 <.0001
Least Squares Means
Standard
Effect diet exertype time Estimate Error DF t Value
diet*exertype 1 1 89.3333 2.3066 24 38.73
diet*exertype 1 2 93.4667 2.3066 24 40.52
diet*exertype 1 3 105.07 2.3066 24 45.55
diet*exertype 2 1 92.3333 2.3066 24 40.03
diet*exertype 2 2 96.9333 2.3066 24 42.03
diet*exertype 2 3 121.07 2.3066 24 52.49
exertype*time 1 1 90.2000 1.8190 48 49.59
exertype*time 1 2 90.9000 2.7114 48 33.53
exertype*time 1 3 91.4000 2.1304 48 42.90
exertype*time 2 1 93.1000 1.8190 48 51.18
exertype*time 2 2 96.6000 2.7114 48 35.63
exertype*time 2 3 95.9000 2.1304 48 45.02
exertype*time 3 1 96.1000 1.8190 48 52.83
exertype*time 3 2 117.10 2.7114 48 43.19
exertype*time 3 3 126.00 2.1304 48 59.14
diet*exertype*time 1 1 1 88.6000 2.5724 48 34.44
diet*exertype*time 1 1 2 89.6000 3.8344 48 23.37
diet*exertype*time 1 1 3 89.8000 3.0128 48 29.81
diet*exertype*time 1 2 1 90.6000 2.5724 48 35.22
diet*exertype*time 1 2 2 94.6000 3.8344 48 24.67
diet*exertype*time 1 2 3 95.2000 3.0128 48 31.60
diet*exertype*time 1 3 1 94.0000 2.5724 48 36.54
diet*exertype*time 1 3 2 109.80 3.8344 48 28.64
diet*exertype*time 1 3 3 111.40 3.0128 48 36.98
diet*exertype*time 2 1 1 91.8000 2.5724 48 35.69
diet*exertype*time 2 1 2 92.2000 3.8344 48 24.05
diet*exertype*time 2 1 3 93.0000 3.0128 48 30.87
diet*exertype*time 2 2 1 95.6000 2.5724 48 37.16
diet*exertype*time 2 2 2 98.6000 3.8344 48 25.71
diet*exertype*time 2 2 3 96.6000 3.0128 48 32.06
diet*exertype*time 2 3 1 98.2000 2.5724 48 38.17
diet*exertype*time 2 3 2 124.40 3.8344 48 32.44
diet*exertype*time 2 3 3 140.60 3.0128 48 46.67
Least Squares Means
Effect diet exertype time Pr > |t|
diet*exertype 1 1 <.0001
diet*exertype 1 2 <.0001
diet*exertype 1 3 <.0001
diet*exertype 2 1 <.0001
diet*exertype 2 2 <.0001
diet*exertype 2 3 <.0001
exertype*time 1 1 <.0001
exertype*time 1 2 <.0001
exertype*time 1 3 <.0001
exertype*time 2 1 <.0001
exertype*time 2 2 <.0001
exertype*time 2 3 <.0001
exertype*time 3 1 <.0001
exertype*time 3 2 <.0001
exertype*time 3 3 <.0001
diet*exertype*time 1 1 1 <.0001
diet*exertype*time 1 1 2 <.0001
diet*exertype*time 1 1 3 <.0001
diet*exertype*time 1 2 1 <.0001
diet*exertype*time 1 2 2 <.0001
diet*exertype*time 1 2 3 <.0001
diet*exertype*time 1 3 1 <.0001
diet*exertype*time 1 3 2 <.0001
diet*exertype*time 1 3 3 <.0001
diet*exertype*time 2 1 1 <.0001
diet*exertype*time 2 1 2 <.0001
diet*exertype*time 2 1 3 <.0001
diet*exertype*time 2 2 1 <.0001
diet*exertype*time 2 2 2 <.0001
diet*exertype*time 2 2 3 <.0001
diet*exertype*time 2 3 1 <.0001
diet*exertype*time 2 3 2 <.0001
diet*exertype*time 2 3 3 <.0001
Tests of Effect Slices
Num Den
Effect diet time DF DF F Value Pr > F
diet*exertype 1 2 24 12.51 0.0002
diet*exertype 2 2 24 44.77 <.0001
exertype*time 1 2 48 2.63 0.0824
exertype*time 2 2 48 25.83 <.0001
exertype*time 3 2 48 77.98 <.0001
diet*exertype*time 1 1 2 48 1.13 0.3326
diet*exertype*time 1 2 2 48 7.53 0.0014
diet*exertype*time 1 3 2 48 13.92 <.0001
diet*exertype*time 2 1 2 48 1.57 0.2194
diet*exertype*time 2 2 2 48 19.76 <.0001
diet*exertype*time 2 3 2 48 77.39 <.0001
From the tests we see that there is a significant difference between the pulse rate over time of the runners of the low fat diet and the runners of the non-low fat diet. The runners of the low fat diet also have significantly different pulse rate from the pulse rate of all the other groups (the runners of the non-low fat diet, the walkers and “rest-ers” of both diet groups). The runners have a different pulse rate over time from the walkers and “rest-ers” combined. The only time we do not have a significant results is when we look at the pulse rate of the walkers and “rest-ers” over time. Here the test has a p-value of 0.0705 which exceeds 0.05 and thus it is not significant.
When looking at diet=1 and diet=2 separately across all time points we find that there is significant differences in the exertype groups. At time=2 and time=3 there is also a significant difference between exertype groups across both diets. There is not a significant differences between the exertype groups when looking at time=1 and diet=1 nor is there a significant differences among the groups when looking at time=1 and diet=2. For all other combinations of diet and time levels there is a significant difference among the exertype groups.
It might be tempting to try and use the same type of estimate statements in proc glm in order to perform similar types of contrasts. Unfortunately, the results of the estimate statement will be for each of the dependent variable rather than across the repeated measure. Thus, it is not possible to test any of the contrasts that we performed in proc mixed in the above analysis in proc glm using an estimate statement since these contrasts are all done across time. In proc glm these contrasts would be performed separately for each time point which is very different from the results we obtained in proc mixed.
Unequally Spaced Time Points
Modeling Time as a Linear Predictor of Pulse
We have another study which is very similar to the one previously discussed except that in this new study the pulse measurements were not taken at regular time points. In this study a baseline pulse measurement was obtained at time = 0 for every individual in the study. However, subsequent pulse measurements were taken at less regular time intervals. The second pulse measurements were taken at approximately 2 minutes (time = 120 seconds); the pulse measurement was obtained at approximately 5 minutes (time = 300 seconds); and the fourth and final pulse measurement was obtained at approximately 10 minutes (time = 600 seconds). The data for this study is displayed below and it is available in the study2 data file.
data study2; input id exertype diet pulse time; cards; 1 1 1 90 0 1 1 1 92 228 1 1 1 93 296 1 1 1 93 639 2 1 1 90 0 2 1 1 92 56 2 1 1 93 434 2 1 1 93 538 3 1 1 97 0 3 1 1 97 150 3 1 1 94 295 3 1 1 94 541 4 1 1 80 0 4 1 1 82 121 4 1 1 83 256 4 1 1 83 575 5 1 1 91 0 5 1 1 92 161 5 1 1 91 252 5 1 1 91 526 6 1 2 83 0 6 1 2 83 73 6 1 2 84 320 6 1 2 84 570 7 1 2 87 0 7 1 2 88 40 7 1 2 90 325 7 1 2 90 730 8 1 2 92 0 8 1 2 94 205 8 1 2 95 276 8 1 2 95 761 9 1 2 97 0 9 1 2 99 57 9 1 2 96 244 9 1 2 96 695 10 1 2 100 0 10 1 2 97 143 10 1 2 100 296 10 1 2 100 722 11 2 1 86 0 11 2 1 86 83 11 2 1 84 262 11 2 1 84 566 12 2 1 93 0 12 2 1 103 116 12 2 1 104 357 12 2 1 104 479 13 2 1 90 0 13 2 1 92 191 13 2 1 93 280 13 2 1 93 709 14 2 1 95 0 14 2 1 96 112 14 2 1 100 219 14 2 1 100 367 15 2 1 89 0 15 2 1 96 96 15 2 1 95 339 15 2 1 95 639 16 2 2 84 0 16 2 2 86 92 16 2 2 89 351 16 2 2 89 508 17 2 2 103 0 17 2 2 109 196 17 2 2 114 213 17 2 2 120 634 18 2 2 92 0 18 2 2 96 117 18 2 2 101 227 18 2 2 101 614 19 2 2 97 0 19 2 2 98 70 19 2 2 100 295 19 2 2 100 515 20 2 2 102 0 20 2 2 104 165 20 2 2 103 302 20 2 2 103 792 21 3 1 93 0 21 3 1 98 100 21 3 1 110 396 21 3 1 115 498 22 3 1 98 0 22 3 1 104 104 22 3 1 112 310 22 3 1 117 518 23 3 1 98 0 23 3 1 105 148 23 3 1 118 208 23 3 1 121 677 24 3 1 87 0 24 3 1 122 171 24 3 1 127 320 24 3 1 133 633 25 3 1 94 0 25 3 1 110 57 25 3 1 116 268 25 3 1 119 657 26 3 2 95 0 26 3 2 126 163 26 3 2 143 382 26 3 2 147 501 27 3 2 100 0 27 3 2 126 70 27 3 2 140 347 27 3 2 148 737 28 3 2 103 0 28 3 2 124 61 28 3 2 140 263 28 3 2 143 588 29 3 2 94 0 29 3 2 135 164 29 3 2 130 353 29 3 2 137 560 30 3 2 99 0 30 3 2 111 114 30 3 2 140 362 30 3 2 148 501 ; run;
In order to get a better understanding of the data we will look at a scatter plot of the data with lines connecting the points for each individual.
proc sort data=study2; by id time; run; goptions reset=all; symbol1 c=blue v=star h=.8 i=j r=10; symbol2 c=red v=dot h=.8 i=j r=10; symbol3 c=green v=square h=.8 i=j r=10; axis1 order=(60 to 150 by 30) label=(a=90 'Pulse'); proc gplot data=study2; plot pulse*time=id / vaxis=axis1; run;
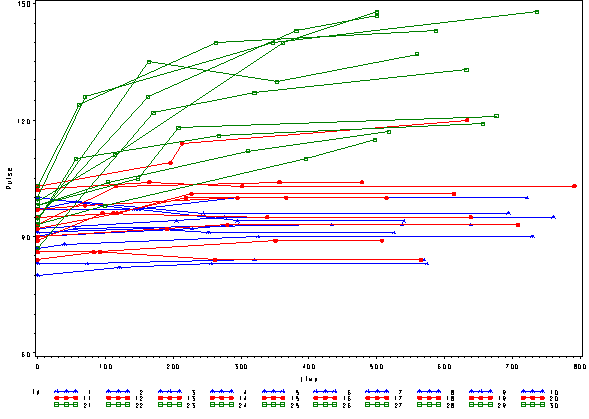
This is a situation where multilevel modeling excels for the analysis of data with irregularly spaced time points. The multilevel model with time as a linear effect is illustrated in the following equations.
Level 1 (time): Pulse = β0j + β1j (Time) + rij Level 2 (person): β0j = γ00 + γ01(Exertype) + u0j Level 2 (person): β1j = γ10 + γ11(Exertype) + u1j
Substituting the level 2 model into the level 1 model we get the following single equations. Note: The random components have been placed in square brackets.
Pulse = γ00 + γ01(Exertype) + γ10(Time) + γ11(Exertype*time) + [ u0j + u1j(Time) + rij ]
Since this model contains both fixed and random components, it can be analyzed in proc mixed as shown below.
*the linear model ; proc mixed data=study2 covtest noclprint; class id exertype ; model pulse = time exertype time*exertype / solution outp=pred1r outpm = pred1f; random intercept time / subject = id; run;
Covariance Parameter Estimates
Standard Z
Cov Parm Subject Estimate Error Value Pr Z
Intercept id 33.8894 13.3635 2.54 0.0056
time id 0.000133 0.000080 1.66 0.0482
Residual 32.4052 5.4327 5.96 <.0001
Solution for Fixed Effects
Standard
Effect exertype Estimate Error DF t Value Pr > |t|
Intercept 103.70 2.2884 27 45.31 <.0001
time 0.05635 0.005405 27 10.43 <.0001
exertype 1 -12.6252 3.2262 60 -3.91 0.0002
exertype 2 -9.1144 3.2309 60 -2.82 0.0065
exertype 3 0 . . . .
time*exertype 1 -0.05477 0.007531 60 -7.27 <.0001
time*exertype 2 -0.04760 0.007711 60 -6.17 <.0001
time*exertype 3 0 . . . .
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
time 1 27 51.13 <.0001
exertype 2 60 8.15 0.0007
time*exertype 2 60 30.68 <.0001
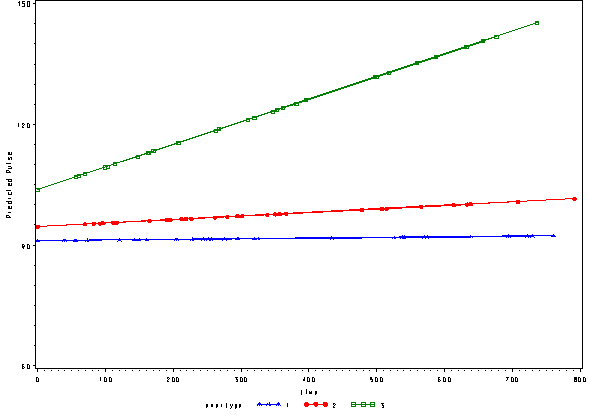
The output file pred1f contains the predicted values based on the fixed part of the model. We can illustrate what the predicted values of pulse look like using this model below.
goptions reset=all; symbol1 c=blue v=star h=.8 i=j; symbol2 c=red v=dot h=.8 i=j; symbol3 c=green v=square h=.8 i=j; axis1 order=(60 to 150 by 30) label=(a=90 'Predicted Pulse'); proc gplot data=pred1f; plot pred*time=exertype /vaxis=axis1; run; quit;
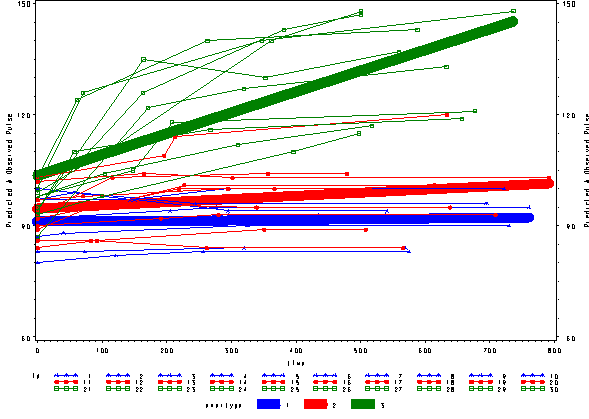
We can include the observed pulse as well and see that this model is not fitting very well at all. The green line is fitting curved data with a straight line.
proc sort data=pred1f; by time; run; goptions reset=all; symbol1 c=blue v=star h=.8 i=j w=10; symbol2 c=red v=dot h=.8 i=j w=10; symbol3 c=green v=square h=.8 i=j w=10; symbol4 c=blue v=star h=.8 i=j r=10; symbol5 c=red v=dot h=.8 i=j r=10; symbol6 c=green v=square h=.8 i=j r=10; axis1 order=(60 to 150 by 30) label=(a=90 'Predicted and Observed Pulse'); proc gplot data=pred1f; plot pred*time=exertype / vaxis=axis1 ; plot2 pulse*time = id / vaxis=axis1 ;; run; quit;
Modeling Time as a Quadratic Predictor of Pulse
To model the quadratic effect of time, we add time*time to the model. We see that term is significant.
*the quadratic model ;
proc mixed data=study2 covtest noclprint;
class id exertype;
model pulse = time exertype time*exertype time*time / solution outp=pred2r outpm=pred2f ;
random intercept time / subject = id;
run;
Covariance Parameter Estimates
Standard Z
Cov Parm Subject Estimate Error Value Pr Z
Intercept id 33.2228 12.3961 2.68 0.0037
time id 0.000151 0.000075 2.00 0.0226
Residual 24.8148 4.2003 5.91 <.0001
Solution for Fixed Effects
Standard
Effect exertype Estimate Error DF t Value Pr > |t|
Intercept 101.68 2.2145 27 45.91 <.0001
time 0.08777 0.008310 27 10.56 <.0001
exertype 1 -12.9233 3.0723 59 -4.21 <.0001
exertype 2 -9.3558 3.0757 59 -3.04 0.0035
exertype 3 0 . . . .
time*exertype 1 -0.05253 0.007332 59 -7.16 <.0001
time*exertype 2 -0.04690 0.007464 59 -6.28 <.0001
time*exertype 3 0 . . . .
time*time -0.00005 0.000011 27 -4.83 <.0001
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
time 1 27 55.38 <.0001
exertype 2 59 9.42 0.0003
time*exertype 2 59 30.48 <.0001
time*time 1 27 23.28 <.0001
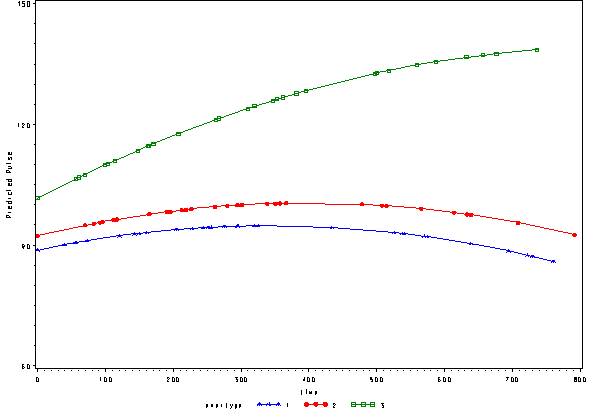
Below we see the predicted values from this model with the quadratic effect of time.
* just predicted, fixed ; proc sort data=pred2f; by time; run; goptions reset=all; symbol1 c=blue v=star h=.8 i=j ; symbol2 c=red v=dot h=.8 i=j ; symbol3 c=green v=square h=.8 i=j ; axis1 order=(60 to 150 by 30) label=(a=90 'Predicted Pulse'); proc gplot data=pred2f; plot pred*time=exertype /vaxis=axis1 ; run; quit;
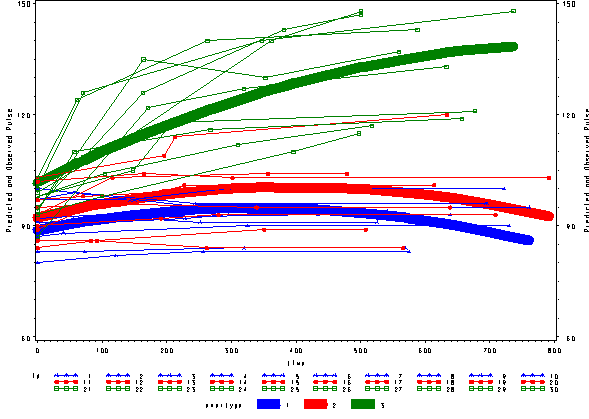
Again, we can plot the predicted values against the actual values of pulse. We see that this model fits better, but it appears that the predicted values for the green group have too little curvature and the red and blue group have too much curvature.
* predicted vs. actual , fixed ; proc sort data=pred2f; by time; run; goptions reset=all; symbol1 c=blue v=star h=.8 i=j w=10; symbol2 c=red v=dot h=.8 i=j w=10; symbol3 c=green v=square h=.8 i=j w=10; symbol4 c=blue v=star h=.8 i=j r=10; symbol5 c=red v=dot h=.8 i=j r=10; symbol6 c=green v=square h=.8 i=j r=10; axis1 order=(60 to 150 by 30) label=(a=90 'Predicted and Observed Pulse'); proc gplot data=pred2f; plot pred*time=exertype / vaxis=axis1 ; plot2 pulse*time = id / vaxis=axis1 ;; run; quit;
Modeling Time as a Quadratic Predictor of Pulse, Interacting by Exertype
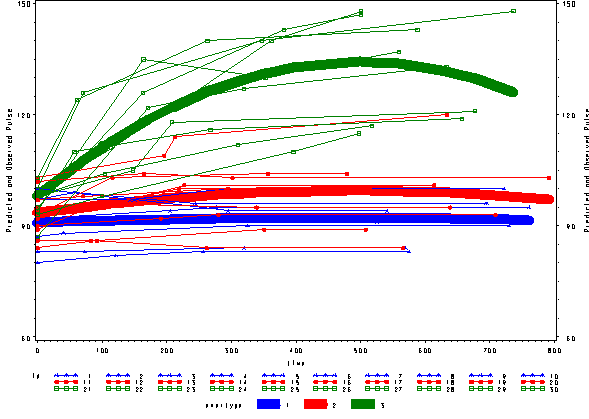
We can include an interaction of time*time*exertype to indicate that the different exercises not only show different linear trends over time, but that they also show different quadratic trends over time, as shown below. The time*time*exertype term is significant.
* quadratic model , model 3 ;
proc mixed data=study2 covtest noclprint;
class id exertype;
model pulse = time exertype time*exertype time*time time*time*exertype / solution outp=pred3r outpm=pred3f ;
random intercept time / subject = id;
run;
Solution for Fixed Effects
Standard
Effect exertype Estimate Error DF t Value Pr > |t|
Intercept 98.0958 2.1923 27 44.75 <.0001
time 0.1448 0.01065 27 13.60 <.0001
exertype 1 -7.2807 3.0989 57 -2.35 0.0223
exertype 2 -4.6201 3.1042 57 -1.49 0.1422
exertype 3 0 . . . .
time*exertype 1 -0.1393 0.01461 57 -9.53 <.0001
time*exertype 2 -0.1204 0.01472 57 -8.18 <.0001
time*exertype 3 0 . . . .
time*time -0.00014 0.000016 27 -9.17 <.0001
time*time*exertype 1 0.000139 0.000021 57 6.67 <.0001
time*time*exertype 2 0.000120 0.000021 57 5.60 <.0001
time*time*exertype 3 0 . . . .
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
time 1 27 96.49 <.0001
exertype 2 57 2.83 0.0676
time*exertype 2 57 52.32 <.0001
time*time 1 27 84.11 <.0001
time*time*exertype 2 57 24.77 <.0001
* predicted vs. actual , fixed ;
proc sort data=pred3f;
by time;
run;
goptions reset=all;
symbol1 c=blue v=star h=.8 i=j w=10;
symbol2 c=red v=dot h=.8 i=j w=10;
symbol3 c=green v=square h=.8 i=j w=10;
symbol4 c=blue v=star h=.8 i=j r=10;
symbol5 c=red v=dot h=.8 i=j r=10;
symbol6 c=green v=square h=.8 i=j r=10;
axis1 order=(60 to 150 by 30) label=(a=90 'Predicted and Observed Pulse');
proc gplot data=pred3f;
plot pred*time=exertype / vaxis=axis1 ;
plot2 pulse*time = id / vaxis=axis1 ;;
run;
quit;
Below we see the predicted and actual values and see that this model fits much better. The green curve hugs the data from the green group much better and the blue and red groups are much flatter, fitting their data much better as well.
For more information