page 150 Table 5.1 Observed (obs) and estimated expected (exp) frequencies within each decile of risk, defined by fitted value (prob.) for dfree = 1 and dfree = 0 using the fitted logistic regression model in Table 4.9.
NOTE: The values in the printout do not match those in the text exactly because Stata and SPSS use different methods of handling ties between values.
Get file='uis.sav'. compute ndrgfp1=1/((ndrugtx+1)/10). compute ndrgfp2=ndrgfp1*ln((ndrugtx+1)/10). compute nage=ndrgfp1*age. compute racesite=race*site. execute. LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /PRINT=GOODFIT.
| Unweighted Cases(a) | N | Percent | |
|---|---|---|---|
| Selected Cases | Included in Analysis | 575 | 100.0 |
| Missing Cases | 0 | .0 | |
| Total | 575 | 100.0 | |
| Unselected Cases | 0 | .0 | |
| Total | 575 | 100.0 | |
| a If weight is in effect, see classification table for the total number of cases. | |||
| Original Value | Internal Value |
|---|---|
| .00 | 0 |
| 1.00 | 1 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 0 | DFREE | .00 | 428 | 0 | 100.0 |
| 1.00 | 147 | 0 | .0 | ||
| Overall Percentage | |
|
74.4 | ||
| a Constant is included in the model. | |||||
| b The cut value is .500 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 0 | Constant | -1.069 | .096 | 124.967 | 1 | .000 | .343 |
| |
Score | df | Sig. | ||
|---|---|---|---|---|---|
| Step 0 | Variables | AGE | 1.406 | 1 | .236 |
| NDRGFP1 | 6.074 | 1 | .014 | ||
| NDRGFP2 | 4.115 | 1 | .043 | ||
| IVHX2 | .207 | 1 | .649 | ||
| IVHX3 | 9.737 | 1 | .002 | ||
| RACE | 4.779 | 1 | .029 | ||
| TREAT | 5.163 | 1 | .023 | ||
| SITE | 1.692 | 1 | .193 | ||
| NAGE | 5.573 | 1 | .018 | ||
| RACESITE | .144 | 1 | .705 | ||
| Overall Statistics | 52.071 | 10 | .000 | ||
| |
Chi-square | df | Sig. | |
|---|---|---|---|---|
| Step 1 | Step | 55.766 | 10 | .000 |
| Block | 55.766 | 10 | .000 | |
| Model | 55.766 | 10 | .000 | |
| Step | -2 Log likelihood | Cox & Snell R Square | Nagelkerke R Square |
|---|---|---|---|
| 1 | 597.963 | .092 | .136 |
| Step | Chi-square | df | Sig. |
|---|---|---|---|
| 1 | 4.419 | 8 | .818 |
| |
DFREE = .00 | DFREE = 1.00 | Total | |||
|---|---|---|---|---|---|---|
| Observed | Expected | Observed | Expected | |||
| Step 1 | 1 | 54 | 53.900 | 4 | 4.100 | 58 |
| 2 | 52 | 51.644 | 6 | 6.356 | 58 | |
| 3 | 51 | 49.425 | 7 | 8.575 | 58 | |
| 4 | 47 | 47.353 | 11 | 10.647 | 58 | |
| 5 | 42 | 45.235 | 16 | 12.765 | 58 | |
| 6 | 46 | 43.163 | 12 | 14.837 | 58 | |
| 7 | 40 | 40.357 | 18 | 17.643 | 58 | |
| 8 | 33 | 37.672 | 25 | 20.328 | 58 | |
| 9 | 37 | 34.282 | 22 | 24.718 | 59 | |
| 10 | 26 | 24.970 | 26 | 27.030 | 52 | |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 1 | DFREE | .00 | 417 | 11 | 97.4 |
| 1.00 | 131 | 16 | 10.9 | ||
| Overall Percentage | |
|
75.3 | ||
| a The cut value is .500 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 1(a) | AGE | .117 | .029 | 16.317 | 1 | .000 | 1.124 |
| NDRGFP1 | 1.669 | .407 | 16.804 | 1 | .000 | 5.307 | |
| NDRGFP2 | .434 | .117 | 13.762 | 1 | .000 | 1.543 | |
| IVHX2 | -.635 | .299 | 4.514 | 1 | .034 | .530 | |
| IVHX3 | -.705 | .262 | 7.263 | 1 | .007 | .494 | |
| RACE | .684 | .264 | 6.708 | 1 | .010 | 1.982 | |
| TREAT | .435 | .204 | 4.556 | 1 | .033 | 1.545 | |
| SITE | .516 | .255 | 4.101 | 1 | .043 | 1.676 | |
| NAGE | -.015 | .006 | 6.419 | 1 | .011 | .985 | |
| RACESITE | -1.429 | .530 | 7.280 | 1 | .007 | .239 | |
| Constant | -6.844 | 1.219 | 31.504 | 1 | .000 | .001 | |
| a Variable(s) entered on step 1: AGE, NDRGFP1, NDRGFP2, IVHX2, IVHX3, RACE, TREAT, SITE, NAGE, RACESITE. | |||||||
page 157 Table 5.2 Classification table based on the logistic regression model in Table 4.9 using a cutpoint of 0.5.
NOTE: The above code gives this table (at step 1).
page 159 Table 5.3 Classification table based on the logistic regression . model in Table 4.9 using a cutpoint of 0.5, but all probabilities pi-hat < 0.5 are replaced with pi-hat = 0.05 and all probabilities pi-hat >= 0.50 are . replaced with pi-hat = 0.95.
NOTE: We were unable to reproduce this table.
page 160 Table 5.4 Classification table based on the logistic regression . model in Table 4.9 using a cutpoint of 0.5, but all probabilities pi-hat < 0.50 are replaced with pi-hat = 0.45 and all probabilities pi-hat >= 0.50 are replaced with pi-hat = 0.55.
NOTE: We were unable to reproduce this table.
page 161 Table 5.5 Classification table based on the logistic regression model in Table 4.9 using a cutpoint of 0.6.
LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /CRITERTIA CUT(.6).
| Unweighted Cases(a) | N | Percent | |
|---|---|---|---|
| Selected Cases | Included in Analysis | 575 | 100.0 |
| Missing Cases | 0 | .0 | |
| Total | 575 | 100.0 | |
| Unselected Cases | 0 | .0 | |
| Total | 575 | 100.0 | |
| a If weight is in effect, see classification table for the total number of cases. | |||
| Original Value | Internal Value |
|---|---|
| .00 | 0 |
| 1.00 | 1 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 0 | DFREE | .00 | 428 | 0 | 100.0 |
| 1.00 | 147 | 0 | .0 | ||
| Overall Percentage | |
|
74.4 | ||
| a Constant is included in the model. | |||||
| b The cut value is .600 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 0 | Constant | -1.069 | .096 | 124.967 | 1 | .000 | .343 |
| |
Score | df | Sig. | ||
|---|---|---|---|---|---|
| Step 0 | Variables | AGE | 1.406 | 1 | .236 |
| NDRGFP1 | 6.074 | 1 | .014 | ||
| NDRGFP2 | 4.115 | 1 | .043 | ||
| IVHX2 | .207 | 1 | .649 | ||
| IVHX3 | 9.737 | 1 | .002 | ||
| RACE | 4.779 | 1 | .029 | ||
| TREAT | 5.163 | 1 | .023 | ||
| SITE | 1.692 | 1 | .193 | ||
| NAGE | 5.573 | 1 | .018 | ||
| RACESITE | .144 | 1 | .705 | ||
| Overall Statistics | 52.071 | 10 | .000 | ||
| |
Chi-square | df | Sig. | |
|---|---|---|---|---|
| Step 1 | Step | 55.766 | 10 | .000 |
| Block | 55.766 | 10 | .000 | |
| Model | 55.766 | 10 | .000 | |
| Step | -2 Log likelihood | Cox & Snell R Square | Nagelkerke R Square |
|---|---|---|---|
| 1 | 597.963 | .092 | .136 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 1 | DFREE | .00 | 428 | 0 | 100.0 |
| 1.00 | 142 | 5 | 3.4 | ||
| Overall Percentage | |
|
75.3 | ||
| a The cut value is .600 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 1(a) | AGE | .117 | .029 | 16.317 | 1 | .000 | 1.124 |
| NDRGFP1 | 1.669 | .407 | 16.804 | 1 | .000 | 5.307 | |
| NDRGFP2 | .434 | .117 | 13.762 | 1 | .000 | 1.543 | |
| IVHX2 | -.635 | .299 | 4.514 | 1 | .034 | .530 | |
| IVHX3 | -.705 | .262 | 7.263 | 1 | .007 | .494 | |
| RACE | .684 | .264 | 6.708 | 1 | .010 | 1.982 | |
| TREAT | .435 | .204 | 4.556 | 1 | .033 | 1.545 | |
| SITE | .516 | .255 | 4.101 | 1 | .043 | 1.676 | |
| NAGE | -.015 | .006 | 6.419 | 1 | .011 | .985 | |
| RACESITE | -1.429 | .530 | 7.280 | 1 | .007 | .239 | |
| Constant | -6.844 | 1.219 | 31.504 | 1 | .000 | .001 | |
| a Variable(s) entered on step 1: AGE, NDRGFP1, NDRGFP2, IVHX2, IVHX3, RACE, TREAT, SITE, NAGE, RACESITE. | |||||||
page 161 Table 5.6 Summary of sensitivity, specificity, and 1-specificity for classification tables based on the logistic regression model in Table 4.9 using a cutpoint of 0.05 to 0.60 in increments of 0.05.
NOTE: We were unable to reproduce this table.
page 162 Figure 5.1 Plot of sensitivity and specificity versus all . possible cutpoints in the UIS.
NOTE: We were unable to reproduce this graph.
page 163 Figure 5.2 Plot of sensitivity versus 1-specificity for all . possible cutpoints in the UIS. The resulting curve is called a ROC curve.
NOTE: We were unable to reproduce this graph.
page 171 Figure 5.3 Plot of leverage (h) versus the estimated logistic probability (pi-hat) for a hypothetical univariable logistic regression model.
NOTE: We cannot recreate this figure because we do not have the hypothetical data that were used.
page 172 Figure 5.4 Plot of the distance portion of leverage (b) versus the estimated logistic probability (pi-hat) for a hypothetical univariable logistic regression model.
NOTE: We cannot recreate this figure because we do not have the hypothetical data that were used.
page 177 Figure 5.5 Plot of delta-chi-square versus the estimated probability from the fitted model in Table 4.9, UIS J = 521 covariate patterns.
NOTE: We have skipped this for now.
page 178 Figure 5.6 Plot of delta-D versus the estimated probability from the fitted model in Table 4.9, UIS J = 521 covariate patterns.
NOTE: We have skipped this for now.
page 179 Figure 5.7 Plot of delta-beta-hat versus the estimated probability from the fitted model in Table 4.9, UIS J = 521 covariate patterns.
LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /SAVE COOK PRED LEV.
| Unweighted Cases(a) | N | Percent | |
|---|---|---|---|
| Selected Cases | Included in Analysis | 575 | 100.0 |
| Missing Cases | 0 | .0 | |
| Total | 575 | 100.0 | |
| Unselected Cases | 0 | .0 | |
| Total | 575 | 100.0 | |
| a If weight is in effect, see classification table for the total number of cases. | |||
| Original Value | Internal Value |
|---|---|
| .00 | 0 |
| 1.00 | 1 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 0 | DFREE | .00 | 428 | 0 | 100.0 |
| 1.00 | 147 | 0 | .0 | ||
| Overall Percentage | |
|
74.4 | ||
| a Constant is included in the model. | |||||
| b The cut value is .500 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 0 | Constant | -1.069 | .096 | 124.967 | 1 | .000 | .343 |
| |
Score | df | Sig. | ||
|---|---|---|---|---|---|
| Step 0 | Variables | AGE | 1.406 | 1 | .236 |
| NDRGFP1 | 6.074 | 1 | .014 | ||
| NDRGFP2 | 4.115 | 1 | .043 | ||
| IVHX2 | .207 | 1 | .649 | ||
| IVHX3 | 9.737 | 1 | .002 | ||
| RACE | 4.779 | 1 | .029 | ||
| TREAT | 5.163 | 1 | .023 | ||
| SITE | 1.692 | 1 | .193 | ||
| NAGE | 5.573 | 1 | .018 | ||
| RACESITE | .144 | 1 | .705 | ||
| Overall Statistics | 52.071 | 10 | .000 | ||
| |
Chi-square | df | Sig. | |
|---|---|---|---|---|
| Step 1 | Step | 55.766 | 10 | .000 |
| Block | 55.766 | 10 | .000 | |
| Model | 55.766 | 10 | .000 | |
| Step | -2 Log likelihood | Cox & Snell R Square | Nagelkerke R Square |
|---|---|---|---|
| 1 | 597.963 | .092 | .136 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 1 | DFREE | .00 | 417 | 11 | 97.4 |
| 1.00 | 131 | 16 | 10.9 | ||
| Overall Percentage | |
|
75.3 | ||
| a The cut value is .500 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 1(a) | AGE | .117 | .029 | 16.317 | 1 | .000 | 1.124 |
| NDRGFP1 | 1.669 | .407 | 16.804 | 1 | .000 | 5.307 | |
| NDRGFP2 | .434 | .117 | 13.762 | 1 | .000 | 1.543 | |
| IVHX2 | -.635 | .299 | 4.514 | 1 | .034 | .530 | |
| IVHX3 | -.705 | .262 | 7.263 | 1 | .007 | .494 | |
| RACE | .684 | .264 | 6.708 | 1 | .010 | 1.982 | |
| TREAT | .435 | .204 | 4.556 | 1 | .033 | 1.545 | |
| SITE | .516 | .255 | 4.101 | 1 | .043 | 1.676 | |
| NAGE | -.015 | .006 | 6.419 | 1 | .011 | .985 | |
| RACESITE | -1.429 | .530 | 7.280 | 1 | .007 | .239 | |
| Constant | -6.844 | 1.219 | 31.504 | 1 | .000 | .001 | |
| a Variable(s) entered on step 1: AGE, NDRGFP1, NDRGFP2, IVHX2, IVHX3, RACE, TREAT, SITE, NAGE, RACESITE. | |||||||
GRAPH /SCATTERPLOT(BIVAR)=pre_1 WITH coo_1.
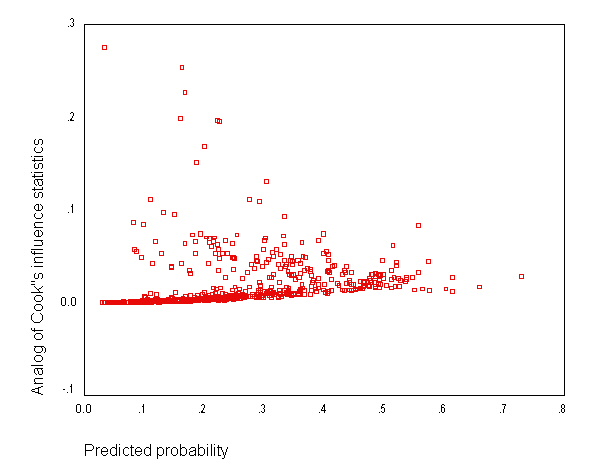
page 180 Figure 5.8 Plot of delta-x-square versus the probability from the fitted model in Table 4.9 with size of the plotting symbol proportional to delta-beta-hat, UIS J = 521 covariate patterns.
NOTE: We have skipped this for now.
page 182 Table 5.8 Covariate values, observed outcome (yj), number (mj), estimated logistic probability (pi-hat), and the value of the four diagnostic statistics delta-beta-hat, delta-x-square, and leverage (h) for the four most. extreme covariate patterns (P#).
SORT CASES BY age (A) ndrgfp1 (A) ndrgfp2 (A) ivhx2 (A) ivhx3 (A) race (A) treat (A) site (A) nage (A) racesite (A) . save outfile = 'd:uissort.sav'. aggregate outfile=* /break=age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /count=n. compute covpat = $casenum. save outfile = 'd:uiscovpat.sav'. match files /file='d:uissort.sav' /table='d:uiscovpat.sav' /by age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite . execute. USE ALL. COMPUTE filter_$=(covpat=31 or covpat=477 or covpat=105 or covpat=468). VARIABLE LABEL filter_$ 'covpat=31 or covpat=477 or covpat=105 or covpat=468 '+ '(FILTER)'. VALUE LABELS filter_$ 0 'Not Selected' 1 'Selected'. FORMAT filter_$ (f1.0). FILTER BY filter_$. EXECUTE . list covpat age ndrugtx ivhx race treat site count pre_1 coo_1 lev_1. The variables are listed in the following order: LINE 1: COVPAT AGE NDRUGTX IVHX RACE TREAT SITE LINE 2: COUNT PRE_1 COO_1 LEV_1 COVPAT: 31.00 24.00 20.00 2.00 .00 .00 1.00 COUNT: 1 .03263 .27429 .00917 COVPAT: 105.00 26.00 .00 1.00 1.00 .00 .00 COUNT: 2 .40300 .05503 .03582 COVPAT: 105.00 26.00 .00 1.00 1.00 .00 .00 COUNT: 2 .40300 .05503 .03582 COVPAT: 468.00 40.00 .00 3.00 1.00 .00 .00 COUNT: 1 .16760 .22610 .04354 COVPAT: 477.00 41.00 .00 3.00 1.00 .00 .00 COUNT: 1 .16263 .25409 .04703 Number of cases read: 5 Number of cases listed: 5
page 183 Table 5.9 Estimated coefficients from all data, the percent change when the covariate pattern is deleted, and values of goodness-of-fit statistics for each model.
USE ALL. COMPUTE filter_$=(covpat ~= 31 and covpat ~= 477 and covpat ~= 105 and covpat ~= 468). VARIABLE LABEL filter_$ 'covpat ~= 31 and covpat ~= 477 and covpat ~= 105 and'+ ' covpat ~= 468 (FILTER)'. VALUE LABELS filter_$ 0 'Not Selected' 1 'Selected'. FORMAT filter_$ (f1.0). FILTER BY filter_$. EXECUTE .
NOTE: This code gives the value for C-hat. The code below gives the coefficients listed in the column labeled "all data".
LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite.
| Unweighted Cases(a) | N | Percent | |
|---|---|---|---|
| Selected Cases | Included in Analysis | 570 | 100.0 |
| Missing Cases | 0 | .0 | |
| Total | 570 | 100.0 | |
| Unselected Cases | 0 | .0 | |
| Total | 570 | 100.0 | |
| a If weight is in effect, see classification table for the total number of cases. | |||
| Original Value | Internal Value |
|---|---|
| .00 | 0 |
| 1.00 | 1 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 0 | DFREE | .00 | 428 | 0 | 100.0 |
| 1.00 | 142 | 0 | .0 | ||
| Overall Percentage | |
|
75.1 | ||
| a Constant is included in the model. | |||||
| b The cut value is .500 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 0 | Constant | -1.103 | .097 | 129.790 | 1 | .000 | .332 |
| |
Score | df | Sig. | ||
|---|---|---|---|---|---|
| Step 0 | Variables | AGE | 1.592 | 1 | .207 |
| NDRGFP1 | 3.827 | 1 | .050 | ||
| NDRGFP2 | 2.118 | 1 | .146 | ||
| IVHX2 | .222 | 1 | .638 | ||
| IVHX3 | 9.886 | 1 | .002 | ||
| RACE | 3.123 | 1 | .077 | ||
| TREAT | 7.094 | 1 | .008 | ||
| SITE | 1.957 | 1 | .162 | ||
| NAGE | 3.288 | 1 | .070 | ||
| RACESITE | .084 | 1 | .772 | ||
| Overall Statistics | 56.859 | 10 | .000 | ||
| |
Chi-square | df | Sig. | |
|---|---|---|---|---|
| Step 1 | Step | 61.628 | 10 | .000 |
| Block | 61.628 | 10 | .000 | |
| Model | 61.628 | 10 | .000 | |
| Step | -2 Log likelihood | Cox & Snell R Square | Nagelkerke R Square |
|---|---|---|---|
| 1 | 578.333 | .102 | .152 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 1 | DFREE | .00 | 417 | 11 | 97.4 |
| 1.00 | 121 | 21 | 14.8 | ||
| Overall Percentage | |
|
76.8 | ||
| a The cut value is .500 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 1(a) | AGE | .138 | .031 | 20.246 | 1 | .000 | 1.147 |
| NDRGFP1 | 2.042 | .443 | 21.259 | 1 | .000 | 7.709 | |
| NDRGFP2 | .525 | .123 | 18.291 | 1 | .000 | 1.691 | |
| IVHX2 | -.702 | .304 | 5.317 | 1 | .021 | .496 | |
| IVHX3 | -.796 | .268 | 8.839 | 1 | .003 | .451 | |
| RACE | .545 | .273 | 3.990 | 1 | .046 | 1.725 | |
| TREAT | .525 | .208 | 6.352 | 1 | .012 | 1.691 | |
| SITE | .504 | .258 | 3.807 | 1 | .051 | 1.656 | |
| NAGE | -.020 | .007 | 9.100 | 1 | .003 | .980 | |
| RACESITE | -1.251 | .539 | 5.393 | 1 | .020 | .286 | |
| Constant | -7.800 | 1.300 | 36.024 | 1 | .000 | .000 | |
| a Variable(s) entered on step 1: AGE, NDRGFP1, NDRGFP2, IVHX2, IVHX3, RACE, TREAT, SITE, NAGE, RACESITE. | |||||||
*Syntax for column 2 of Table 5.9. temporary. select if (covpat ne 31). LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /print= goodfit. *Syntax for column 3 of Table 5.9. temporary. select if (covpat ne 477). LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /print= goodfit.
*Syntax for column 4 of Table 5.9. temporary. select if (covpat ne 105). LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /print= goodfit. *Syntax for column 5 of Table 5.9. temporary. select if (covpat ne 468). LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /print= goodfit. *Syntax for column 6 of Table 5.9. temporary. select if (covpat ne 31 or covpat ne 477 or covpat ne 105 or covpat ne 468 ). LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite /print= goodfit.
page 189 Table 5.10 Estimated coefficients, standard errors, z-scores, two-tailed p-values and 95% confidence intervals for the final logistic regression model for the UIS (n=575).
LOGISTIC REGRESSION VAR=dfree /METHOD=ENTER age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site nage racesite.
| Unweighted Cases(a) | N | Percent | |
|---|---|---|---|
| Selected Cases | Included in Analysis | 570 | 100.0 |
| Missing Cases | 0 | .0 | |
| Total | 570 | 100.0 | |
| Unselected Cases | 0 | .0 | |
| Total | 570 | 100.0 | |
| a If weight is in effect, see classification table for the total number of cases. | |||
| Original Value | Internal Value |
|---|---|
| .00 | 0 |
| 1.00 | 1 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 0 | DFREE | .00 | 428 | 0 | 100.0 |
| 1.00 | 142 | 0 | .0 | ||
| Overall Percentage | |
|
75.1 | ||
| a Constant is included in the model. | |||||
| b The cut value is .500 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 0 | Constant | -1.103 | .097 | 129.790 | 1 | .000 | .332 |
| |
Score | df | Sig. | ||
|---|---|---|---|---|---|
| Step 0 | Variables | AGE | 1.592 | 1 | .207 |
| NDRGFP1 | 3.827 | 1 | .050 | ||
| NDRGFP2 | 2.118 | 1 | .146 | ||
| IVHX2 | .222 | 1 | .638 | ||
| IVHX3 | 9.886 | 1 | .002 | ||
| RACE | 3.123 | 1 | .077 | ||
| TREAT | 7.094 | 1 | .008 | ||
| SITE | 1.957 | 1 | .162 | ||
| NAGE | 3.288 | 1 | .070 | ||
| RACESITE | .084 | 1 | .772 | ||
| Overall Statistics | 56.859 | 10 | .000 | ||
| |
Chi-square | df | Sig. | |
|---|---|---|---|---|
| Step 1 | Step | 61.628 | 10 | .000 |
| Block | 61.628 | 10 | .000 | |
| Model | 61.628 | 10 | .000 | |
| Step | -2 Log likelihood | Cox & Snell R Square | Nagelkerke R Square |
|---|---|---|---|
| 1 | 578.333 | .102 | .152 |
| |
Predicted | ||||
|---|---|---|---|---|---|
| DFREE | Percentage Correct | ||||
| Observed | .00 | 1.00 | |||
| Step 1 | DFREE | .00 | 417 | 11 | 97.4 |
| 1.00 | 121 | 21 | 14.8 | ||
| Overall Percentage | |
|
76.8 | ||
| a The cut value is .500 | |||||
| |
B | S.E. | Wald | df | Sig. | Exp(B) | |
|---|---|---|---|---|---|---|---|
| Step 1(a) | AGE | .138 | .031 | 20.246 | 1 | .000 | 1.147 |
| NDRGFP1 | 2.042 | .443 | 21.259 | 1 | .000 | 7.709 | |
| NDRGFP2 | .525 | .123 | 18.291 | 1 | .000 | 1.691 | |
| IVHX2 | -.702 | .304 | 5.317 | 1 | .021 | .496 | |
| IVHX3 | -.796 | .268 | 8.839 | 1 | .003 | .451 | |
| RACE | .545 | .273 | 3.990 | 1 | .046 | 1.725 | |
| TREAT | .525 | .208 | 6.352 | 1 | .012 | 1.691 | |
| SITE | .504 | .258 | 3.807 | 1 | .051 | 1.656 | |
| NAGE | -.020 | .007 | 9.100 | 1 | .003 | .980 | |
| RACESITE | -1.251 | .539 | 5.393 | 1 | .020 | .286 | |
| Constant | -7.800 | 1.300 | 36.024 | 1 | .000 | .000 | |
| a Variable(s) entered on step 1: AGE, NDRGFP1, NDRGFP2, IVHX2, IVHX3, RACE, TREAT, SITE, NAGE, RACESITE. | |||||||
page 190 Table 5.11 Estimated odds ratios and 95% confidence intervals for treatment and history of IV drug use in the UIS (N = 575).
NOTE: The code above also gives the values for this table.
page 192 Table 5.12 Estimated odds ratios and 95% confidence intervals for race within site in the UIS (n = 575).
NOTE: We were unable to reproduce these values using SPSS.
page 194 Figure 5.9 Estimated odds ratio and 95% confidence limits for a five-year increase in age based on the model in Table 5.10.
NOTE: We were unable to reproduce this graph.
page 197 Figure 5.10 Estimated odds ratios and 95% confidence limits for an increase of one drug treatment from the plotted value of NDRGTX for a subject of age (a) 20, (b) 25, (c) 30 and (d) 35.
NOTE: We were unable to reproduce this graph.
page 199 Figure 5.11 Estimated odds ratios and 95% confidence limits . comparing zero, two, three up to 10 previous drug treatments to one previous treatment for a subject of age (a) 20, (b) 25, (c) 30 and (d) 35.
NOTE: We were unable to reproduce this graph.
