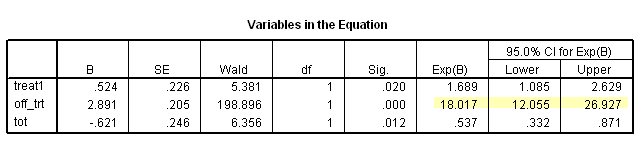The whas500, actg320, gbcs, uis and grace1000 data sets are used in this chapter.
Table 7.1 on page 211 using the actg320 data set.
get file ='C:Dataasa2actg320.sav'. examine variables = cd4 /plot none /percentiles(25,50,75). if cd4<=23 cd4_q = 1. if cd4>23 & cd4<=74.5 cd4_q =2. if cd4>74.5 & cd4<=136.5 cd4_q = 3. if cd4>136.5 cd4_q=4. exe. compute ivdrug_d= (ivdrug>1). compute karnof_90 = (karnof=90). compute karnof_70_80 = (karnof=70|karnof=80). exe. coxreg time with tx ivdrug_d karnof_90 karnof_70_80 age /status=censor(1) /method = enter tx ivdrug_d karnof_90 karnof_70_80 age /strata = cd4_q.
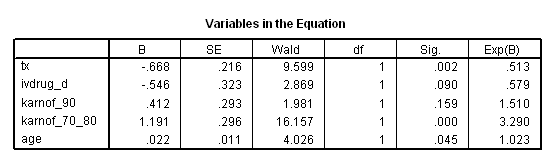
Figure 7.1 on page 212 using the same model from previous example.
coxreg time with tx ivdrug_d karnof_90 karnof_70_80 age
/status=censor(1)
/method = enter tx ivdrug_d karnof_90 karnof_70_80 age
/strata = cd4_q
/save Hazard(H).
* the difference is due to rounding error.
compute xbeta = tx* (-.6678 )+ ivdrug_d *(-.5463) + karnof_90*.412
+ karnof_70_80 *1.191 + age*.0225.
compute rm = xbeta - tx*(-0.6678).
exe.
examine variables =rm by cd4_q
/percentiles(50)
/plot none
/statistics none.
compute s0 = exp(-H/exp(xbeta)). exe. if cd4_q=1 s0_1 = s0**exp(1.2188). if cd4_q=1 s0_1_tx = s0**exp(1.2188-.6678). if cd4_q=2 s0_2 = s0**exp(1.177). if cd4_q=2 s0_2_tx = s0**exp(1.177 - .6678). if cd4_q=3 s0_3 = s0**exp(1.087). if cd4_q=3 s0_3_tx = s0**exp(1.087-.6678). if cd4_q=4 s0_4 = s0**exp(1.0947). if cd4_q=4 s0_4_tx = s0**exp(1.0947-.6678). exe. graph /scatterplot(overlay)=time time with s0_1 s0_1_tx (pair). graph /scatterplot(overlay)=time time with s0_2 s0_2_tx (pair). graph /scatterplot(overlay)=time time with s0_3 s0_3_tx (pair). graph /scatterplot(overlay)=time time with s0_4 s0_4_tx (pair).
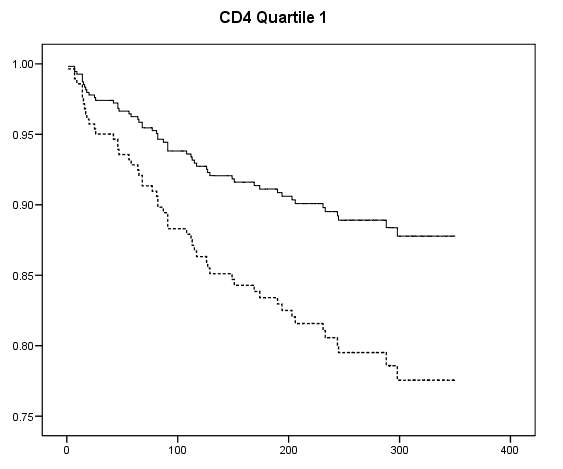
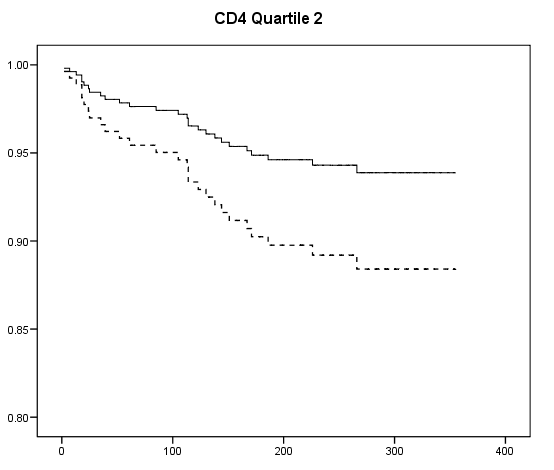
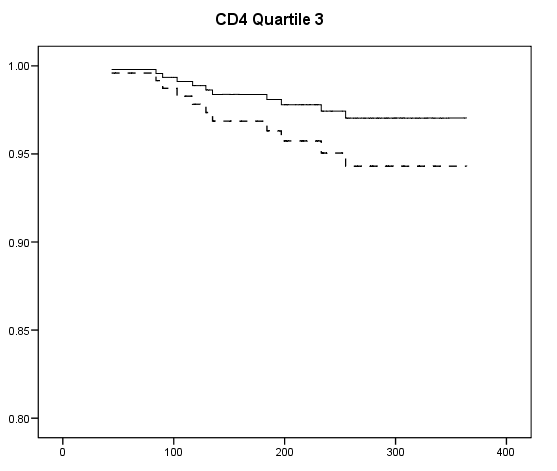
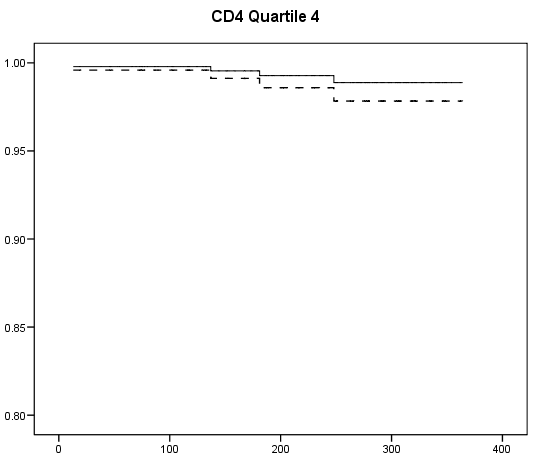
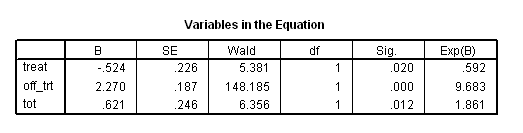
Table 7.2 on page 217 and Table 7.3 on page 219 using the uis data. Due to the lack of post-estimation facility, we will run three models using different coding scheme to achieve the same estimates seen in Table 7.3.
get file ='C:Dataasa2uis.sav'.
* Table 7.2. time program. if T_ <=los off_trt = 0. if T_ > los off_trt = 1. compute tot = treat*off_trt. coxreg time with treat off_trt tot /status censor(1) /method = enter treat off_trt tot.
* Table 7.3, lines 1 and 3. time program. if T_ <=los off_trt = 0. if T_ > los off_trt = 1. compute tot = treat*off_trt. coxreg time with treat off_trt tot /status censor(1) /method = enter treat off_trt tot /print =ci.* Table 7.3, line 2. time program. if T_ <=los off_trt = 1. if T_ > los off_trt = 0. compute tot = treat*off_trt. coxreg time with treat off_trt tot /status censor(1) /method = enter treat off_trt tot /print =ci.
* Table 7.3, line 4. compute treat1= 1-treat. time program. if T_ <=los off_trt = 0. if T_ > los off_trt = 1. compute tot = treat1*off_trt. coxreg time with treat1 off_trt tot /status censor(1) /method = enter treat1 off_trt tot /print =ci.
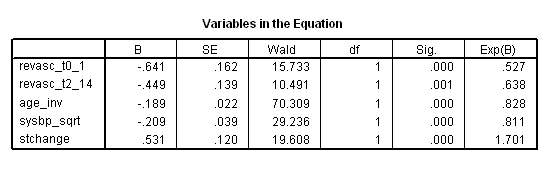
Table 7.5 on page 222 and Table 7.6 on page 223 using the grace1000 data.
get file ='C:Dataasa2grace1000.sav'. compute age_inv = 1/age*1000. compute sysbp_sqrt = sqrt(sysbp). exe. * Table 7.5. coxreg days with revasc age_inv sysbp_sqrt stchange /status death(1) /method = enter revasc age_inv sysbp_sqrt stchange.
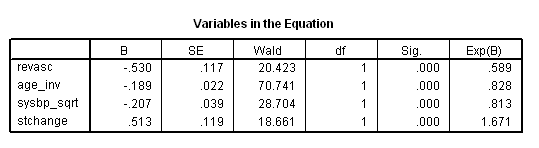
* Table 7.6. time program. if T_ <=revascdays | revasc = 0 revasc_t = 0. if T_ > revascdays & revasc = 1 revasc_t = 1. coxreg days with revasc_t age_inv sysbp_sqrt stchange /status death(1) /method = enter revasc_t age_inv sysbp_sqrt stchange.
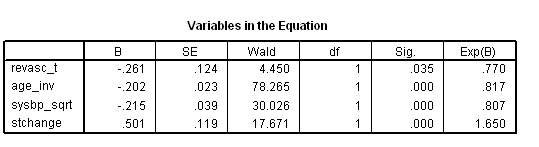
Table 7.7 on page 223 continuing to use the grace1000 data.
use all. compute filter_$ = (revasc=1). filter by filter_$. exe. frequencies variables=revascdays.
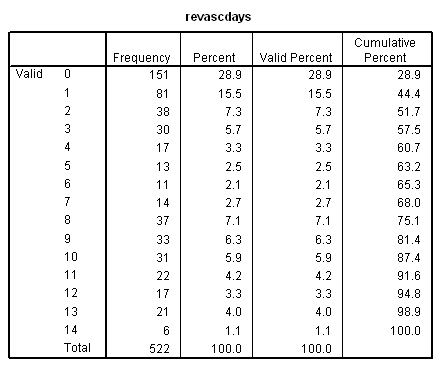
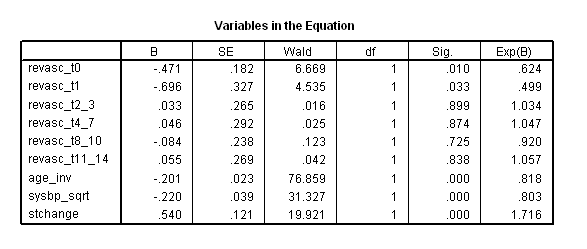
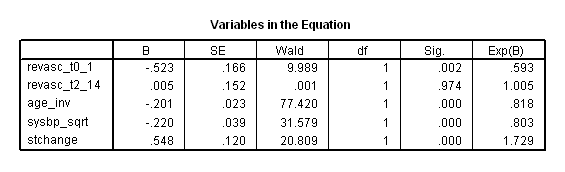
Table 7.8, Table 7.9 and Table 7.11 on page 225 and page 226 using grace1000 data. Notice that the result here does not match with the result in the book
get file ='C:Dataasa2grace1000.sav'. compute age_inv = 1/age*1000. compute sysbp_sqrt = sqrt(sysbp). exe.
time program. compute revasc_t0 = 0. if (T_ > revascdays & revascdays = 0 & revasc=1) revasc_t0 = 1. compute revasc_t1 = 0. if (T_ > revascdays & revascdays = 1 & revasc=1) revasc_t1 = 1. compute revasc_t2_3 = 0. if (T_ > revascdays & revascdays>=2 & revascdays <=3 & revasc=1) revasc_t2_3 = 1. compute revasc_t4_7 = 0. if (T_ > revascdays & revascdays >=4 & revascdays <=7 & revasc=1) revasc_t4_7 = 1. compute revasc_t8_10 = 0. if (T_ > revascdays & revascdays>=8 & revascdays <=10 & revasc=1) revasc_t8_10 = 1. compute revasc_t11_14 = 0. if (T_ > revascdays & revascdays>=11 & revascdays <=14 & revasc=1) revasc_t11_14 = 1. coxreg days with revasc_t0 revasc_t1 revasc_t2_3 revasc_t4_7 revasc_t8_10 revasc_t11_14 age_inv sysbp_sqrt stchange /status death(1) /method = enter revasc_t0 revasc_t1 revasc_t2_3 revasc_t4_7 revasc_t8_10 revasc_t11_14 age_inv sysbp_sqrt stchange .
*table 7_9. time program. compute revasc_t0_1 = 0. if (T_ > revascdays & (revascdays >= 0 & revascdays<=1) & revasc=1) revasc_t0_1 = 1. compute revasc_t2_14 = 0. if (T_ > revascdays & (revascdays >= 2 & revascdays<=14) & revasc=1) revasc_t2_14= 1. coxreg days with revasc_t0_1 revasc_t2_14 age_inv sysbp_sqrt stchange /status death(1) /method = enter revasc_t0_1 revasc_t2_14 age_inv sysbp_sqrt stchange .
time program. compute revasc_t0_1 = 0. compute revasc_t2_14 = 0. if (revasc=0 | (revasc=1 & revascdays>=15) revasc_t0_1 =0. if (revasc=0 | (revasc=1 & revascdays>=15) revasc_t2_14 =0. if (revasc=1 & revascdays<2) revasc_t0_1 =1. if (revasc=1 & revascdays<2) revasc_t2_14 =0. if (revasc=1 & (revascdays>=2 & revascdays<=14)) revasc_t0_1 =0. if (revasc=1 & (revascdays>=2 & revascdays<=14)) revasc_t2_14 =1. coxreg days with revasc_t0_1 revasc_t2_14 age_inv sysbp_sqrt stchange /status death(1) /method = enter revasc_t0_1 revasc_t2_14 age_inv sysbp_sqrt stchange .
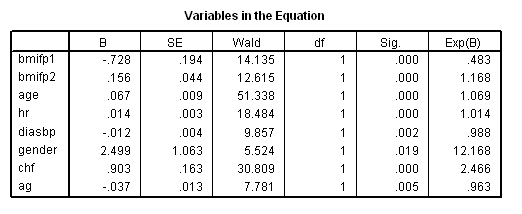
Table 7.12 on page 229 using the whas500 data.
get file ='C:Dataasa2whas500.sav'. use all. compute filter=(dstat = 0 and lenfol > los and id ~=416). filter by filter. exe. compute bmifp1=(bmi/10)**2. compute bmifp2=(bmi/10)**3. compute ag = age*gender. exe. coxreg lenfol with bmifp1 bmifp2 age hr diasbp gender chf ag /status=fstat(1) /method = enter bmifp1 bmifp2 age hr diasbp gender chf ag.
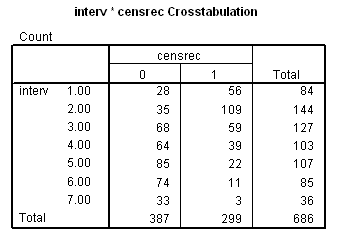
Table 7.13 on page 235 using the gbcs data.
get file ='C:Dataasa2gbcs.sav'. compute month = 12*rectime/365.25. exe. recode month (0 thru 12=1) (12 thru 24=2) (24 thru 36=3) (36 thru 48=4) (48 thru 60=5) (60 thru 72=6) (72 thru Highest=7) into interv . exe. crosstabs /tables =interv by censrec.
Table 7.14 on page 237 using the gbcs data. We need to expand the data for discrete survival analysis.
numeric counter (F3). compute z = 0. loop counter = 1 to interv. if (counter = interv) z = censrec. xsave outfile=expand. end loop. exe. get file= expand. compute month = interv*12. exe. use all. compute tab7_14 = (id = 230 | id = 621 | id = 17 | id =321 | id = 117 |id = 288 | id =278). filter by tab7_14. list variables = id month counter censrec z age.
id month counter censrec z age 17 24.00 1 1 .00 62 17 24.00 2 1 1.00 62 117 36.00 1 1 .00 65 117 36.00 2 1 .00 65 117 36.00 3 1 1.00 65 230 12.00 1 1 1.00 46 278 60.00 1 1 .00 70 278 60.00 2 1 .00 70 278 60.00 3 1 .00 70 278 60.00 4 1 .00 70 278 60.00 5 1 1.00 70 288 36.00 1 0 .00 57 288 36.00 2 0 .00 57 288 36.00 3 0 .00 57 321 24.00 1 0 .00 43 321 24.00 2 0 .00 43 621 12.00 1 0 .00 65
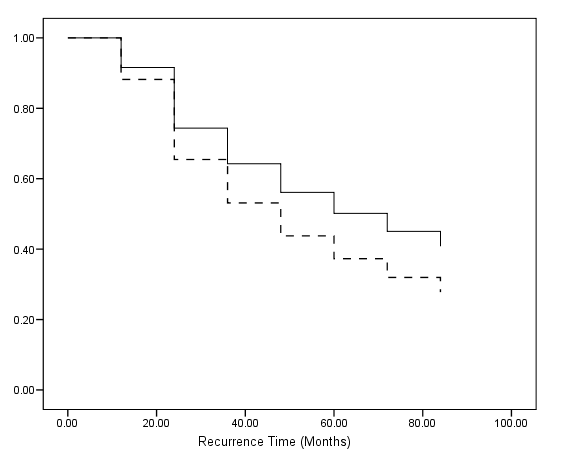
Table 7.15 on page 237 and Figure 7.2 on page 240 using the data created from previous example.
use all. genlin z (reference=0) by counter with age hormone size /model counter age hormone size intercept=no distribution = binomial link = cloglog.
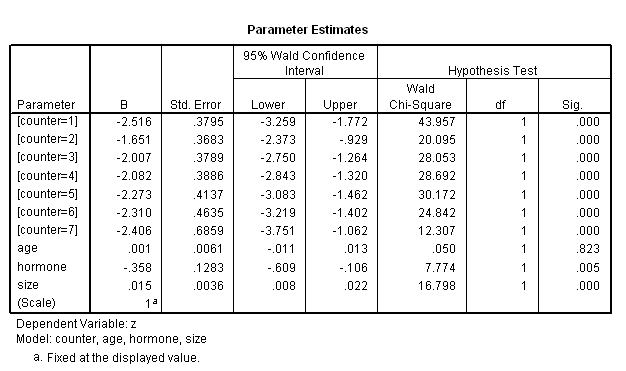
genlin z (reference=0) by counter with age hormone size /model counter age hormone size intercept=no distribution = binomial link = cloglog /outfile covb = "c:/data/asa2/parm_t7_15.sav". get file ='C:Dataasa2parm_t7_15.sav'. compute p0=0. exe. use all. select if (RowType_="EST"). exe. varstocases /make effect from p0 p1 to p7 /index = time. if (time=1) s0 = 1. if $casenum>1 s0 = lag(s0)*exp(-exp(effect)). exe. compute s_nh = s0**(exp(53*p8+25*p10)). compute s_h = s0**(exp(53*p8+ p9 + 25*p10)). exe. compute month = (time-1)*12. exe.
graph /scatterplot(overlay)=month month with s_h s_nh (pair).