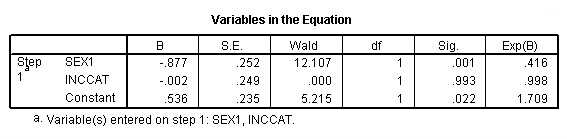
Page 412 Table 17.1
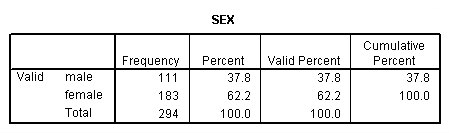
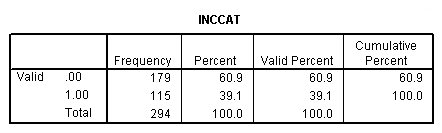
get file 'c:cama3depress.sav'. compute inccat = 0. if income ge 20 inccat = 1. crosstabs sex by inccat.
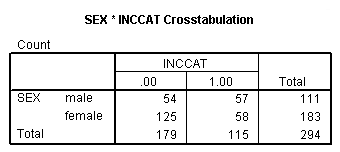
Page 413 Table 17.2
crosstabs sex by inccat by treat.
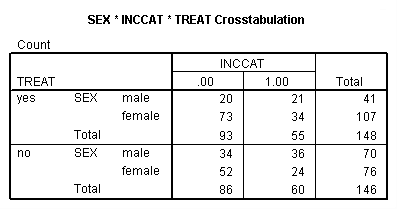
Page 414 Table 17.3
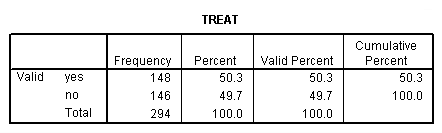
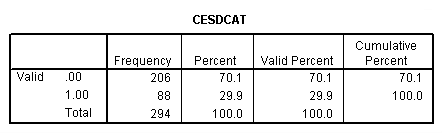
compute cesdcat = 0. if cesd ge 11 cesdcat = 1. crosstab sex by inccat by treat by cesdcat. frequencies var = treat cesdcat sex inccat.
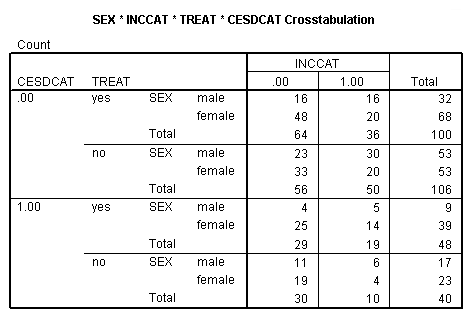
Page 416 middle of the page
crosstabs sex by inccat /cells = expected.
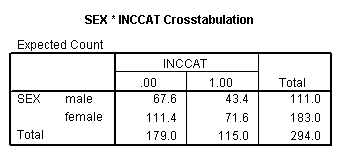
Page 419 middle of the page
crosstabs sex by inccat /statistics = chisq.
<some output omitted>
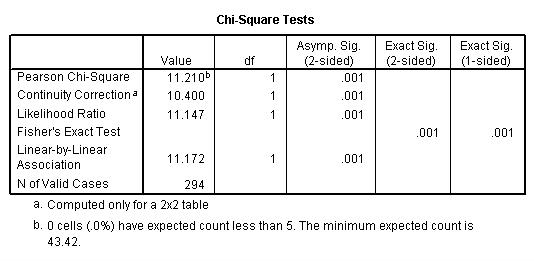
Page 420 Table 17.7
if (sex=1) sex1=-1. if (sex=2) sex1 =1. if (inccat=0) inccat1=-1. if (inccat=1) inccat1 =1. compute sexinc = sex1*inccat1. execute. genlog sex inccat with sex1 inccat1 sexinc /model = poisson /print = estim /plot = none /design inccat1 sex1 sexinc.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
GENERAL LOGLINEAR ANALYSIS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Data Information
294 cases are accepted.
0 cases are rejected because of missing data.
294 weighted cases will be used in the analysis.
4 cells are defined.
0 structural zeros are imposed by design.
0 sampling zeros are encountered.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Variable Information
Factor Levels Value
SEX 2
1.00 male
2.00 female
INCCAT 2
.00
1.00
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Covariates
SEX1
INCCAT1
SEXINC
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Model and Design Information
Model: Poisson
Design: Constant + INCCAT1 + SEX1 + SEXINC
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Correspondence Between Parameters and Terms of the Design
Parameter Aliased Term
1 Constant
2 INCCAT1
3 SEX1
4 SEXINC
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
GENERAL LOGLINEAR ANALYSIS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Convergence Information
Maximum number of iterations: 20
Relative difference tolerance: .001
Final relative difference: 9.12709E-14
Maximum likelihood estimation converged at iteration 1.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Goodness-of-fit Statistics
Chi-Square DF Sig.
Likelihood Ratio .0000 0 .
Pearson .0000 0 .
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Parameter Estimates
Asymptotic 95% CI
Parameter Estimate SE Z-value Lower Upper
1 4.2378 .0616 68.75 4.12 4.36
2 -.1774 .0616 -2.88 -.30 -.06
3 .2128 .0616 3.45 .09 .33
4 -.2042 .0616 -3.31 -.33 -.08
Page 427
Row 3 comparing all two-factor association and first order terms with saturated model
genlog sex treat inccat /model = poisson /print=none /plot=none /design inccat*sex inccat*treat sex*treat.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
GENERAL LOGLINEAR ANALYSIS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Data Information
294 cases are accepted.
0 cases are rejected because of missing data.
294 weighted cases will be used in the analysis.
8 cells are defined.
0 structural zeros are imposed by design.
0 sampling zeros are encountered.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Variable Information
Factor Levels Value
SEX 2
1.00 male
2.00 female
TREAT 2 Has a doctor prescribed or recommended that you take
1.00 yes
2.00 no
INCCAT 2
.00
1.00
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Model and Design Information
Model: Poisson
Design: Constant + SEX*INCCAT + TREAT*INCCAT + SEX*TREAT
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Correspondence Between Parameters and Terms of the Design
Parameter Aliased Term
1 Constant
2 [SEX = 1.00]*[INCCAT = .00]
3 [SEX = 1.00]*[INCCAT = 1.00]
4 [SEX = 2.00]*[INCCAT = .00]
5 x [SEX = 2.00]*[INCCAT = 1.00]
6 [TREAT = 1.00]*[INCCAT = .00]
7 [TREAT = 1.00]*[INCCAT = 1.00]
8 x [TREAT = 2.00]*[INCCAT = .00]
9 x [TREAT = 2.00]*[INCCAT = 1.00]
10 [SEX = 1.00]*[TREAT = 1.00]
11 x [SEX = 1.00]*[TREAT = 2.00]
12 x [SEX = 2.00]*[TREAT = 1.00]
Parameter Aliased Term
13 x [SEX = 2.00]*[TREAT = 2.00]
Note: 'x' indicates an aliased (or a redundant) parameter.
These parameters are set to zero.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Convergence Information
Maximum number of iterations: 20
Relative difference tolerance: .001
Final relative difference: .0003
Maximum likelihood estimation converged at iteration 2.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Goodness-of-fit Statistics
Chi-Square DF Sig.
Likelihood Ratio .0012 1 .9726
Pearson .0012 1 .9726
Row 2 comparing all first order terms with saturated
genlog sex treat inccat /model=poisson /print=none /plot=none /design inccat sex treat.
<some output omitted>
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Model and Design Information
Model: Poisson
Design: Constant + INCCAT + SEX + TREAT
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Correspondence Between Parameters and Terms of the Design
Parameter Aliased Term
1 Constant
2 [INCCAT = .00]
3 x [INCCAT = 1.00]
4 [SEX = 1.00]
5 x [SEX = 2.00]
6 [TREAT = 1.00]
7 x [TREAT = 2.00]
Note: 'x' indicates an aliased (or a redundant) parameter.
These parameters are set to zero.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
GENERAL LOGLINEAR ANALYSIS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Convergence Information
Maximum number of iterations: 20
Relative difference tolerance: .001
Final relative difference: 2.60437E-06
Maximum likelihood estimation converged at iteration 4.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Goodness-of-fit Statistics
Chi-Square DF Sig.
Likelihood Ratio 24.0763 4 8.E-05
Pearson 24.6519 4 6.E-05
Row 1 comparing the empty model with the model of all two-factor models
To obtain the comparison with row 2 of the table, subtract the value of row 2 from the model below. Note also that the degrees of freedom listed in the output is incorrect, showing one more than it should. This is because we had to add the extra constant term to the model to get it to run. (We were unable to figure out how to get SPSS genlog to run a constant only model, so we created a constant and used that in addition to the one SPSS used.)
compute cons = 0. exe. genlog sex treat inccat with cons /model = poisson /print = none /plot = none /design cons.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Model and Design Information
Model: Poisson
Design: Constant + CONS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Correspondence Between Parameters and Terms of the Design
Parameter Aliased Term
1 Constant
2 x CONS
Note: 'x' indicates an aliased (or a redundant) parameter.
These parameters are set to zero.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
GENERAL LOGLINEAR ANALYSIS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Convergence Information
Maximum number of iterations: 20
Relative difference tolerance: .001
Final relative difference: 8.68524E-06
Maximum likelihood estimation converged at iteration 4.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Goodness-of-fit Statistics
Chi-Square DF Sig.
Likelihood Ratio 55.9474 7 1.E-09
Pearson 61.3197 7 8.E-11
Page 428
hiloglinear sex(1 2) treat(1 2) inccat(0 1) /method=backward /print=none /design inccat*sex inccat*treat sex*treat.
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
DATA Information
294 unweighted cases accepted.
0 cases rejected because of out-of-range factor values.
0 cases rejected because of missing data.
294 weighted cases will be used in the analysis.
FACTOR Information
Factor Level Label
SEX 2
TREAT 2 Has a doctor prescribed or recom
INCCAT 2
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
Backward Elimination (p = .050) for DESIGN 1 with generating class
INCCAT*SEX
INCCAT*TREAT
SEX*TREAT
Likelihood ratio chi square = .00118 DF = 1 P = .973
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
If Deleted Simple Effect is DF L.R. Chisq Change Prob Iter
INCCAT*SEX 1 10.669 .0011 2
INCCAT*TREAT 1 .000 .9934 2
SEX*TREAT 1 12.451 .0004 2
Step 1
The best model has generating class
INCCAT*SEX
SEX*TREAT
Likelihood ratio chi square = .00125 DF = 2 P = .999
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
If Deleted Simple Effect is DF L.R. Chisq Change Prob Iter
INCCAT*SEX 1 11.147 .0008 2
SEX*TREAT 1 12.928 .0003 2
Step 2
The best model has generating class
INCCAT*SEX
SEX*TREAT
Likelihood ratio chi square = .00125 DF = 2 P = .999
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
The final model has generating class
INCCAT*SEX
SEX*TREAT
The Iterative Proportional Fit algorithm converged at iteration 0.
The maximum difference between observed and fitted marginal totals is .000
and the convergence criterion is .250
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Goodness-of-fit test statistics
Likelihood ratio chi square = .00125 DF = 2 P = .999
Pearson chi square = .00125 DF = 2 P = .999
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Page 429 The description of the stepwise analysis in the second half of the page.
hiloglinear inccat (0 1) sex1 (0 1) treat1 (0 1) cesdcat (0 1) /method = backward /maxorder=3 /design = inccat*sex1*treat1*cesdcat.
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
DATA Information
294 unweighted cases accepted.
0 cases rejected because of out-of-range factor values.
0 cases rejected because of missing data.
294 weighted cases will be used in the analysis.
FACTOR Information
Factor Level Label
INCCAT 2
SEX1 2
TREAT1 2
CESDCAT 2
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
Backward Elimination (p = .050) for DESIGN 1 with generating class
INCCAT*SEX1*TREAT1
INCCAT*SEX1*CESDCAT
INCCAT*TREAT1*CESDCAT
SEX1*TREAT1*CESDCAT
Likelihood ratio chi square = .04568 DF = 1 P = .831
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
If Deleted Simple Effect is DF L.R. Chisq Change Prob Iter
INCCAT*SEX1*TREAT1 1 .008 .9294 4
INCCAT*SEX1*CESDCAT 1 .009 .9225 3
INCCAT*TREAT1*CESDCAT 1 4.742 .0294 2
SEX1*TREAT1*CESDCAT 1 1.227 .2681 3
Step 1
The best model has generating class
INCCAT*SEX1*CESDCAT
INCCAT*TREAT1*CESDCAT
SEX1*TREAT1*CESDCAT
Likelihood ratio chi square = .05353 DF = 2 P = .974
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
If Deleted Simple Effect is DF L.R. Chisq Change Prob Iter
INCCAT*SEX1*CESDCAT 1 .011 .9173 3
INCCAT*TREAT1*CESDCAT 1 4.740 .0295 2
SEX1*TREAT1*CESDCAT 1 1.241 .2653 4
Step 2
The best model has generating class
INCCAT*TREAT1*CESDCAT
SEX1*TREAT1*CESDCAT
INCCAT*SEX1
Likelihood ratio chi square = .06431 DF = 3 P = .996
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
If Deleted Simple Effect is DF L.R. Chisq Change Prob Iter
INCCAT*TREAT1*CESDCAT 1 4.986 .0256 3
SEX1*TREAT1*CESDCAT 1 1.239 .2656 3
INCCAT*SEX1 1 10.495 .0012 2
Step 3
The best model has generating class
INCCAT*TREAT1*CESDCAT
INCCAT*SEX1
SEX1*TREAT1
SEX1*CESDCAT
Likelihood ratio chi square = 1.30361 DF = 4 P = .861
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
If Deleted Simple Effect is DF L.R. Chisq Change Prob Iter
INCCAT*TREAT1*CESDCAT 1 4.273 .0387 3
INCCAT*SEX1 1 9.783 .0018 3
SEX1*TREAT1 1 11.853 .0006 3
SEX1*CESDCAT 1 2.218 .1364 3
Step 4
The best model has generating class
INCCAT*TREAT1*CESDCAT
INCCAT*SEX1
SEX1*TREAT1
Likelihood ratio chi square = 3.52193 DF = 5 P = .620
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
If Deleted Simple Effect is DF L.R. Chisq Change Prob Iter
INCCAT*TREAT1*CESDCAT 1 4.397 .0360 3
INCCAT*SEX1 1 10.669 .0011 2
SEX1*TREAT1 1 12.451 .0004 2
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
Step 5
The best model has generating class
INCCAT*TREAT1*CESDCAT
INCCAT*SEX1
SEX1*TREAT1
Likelihood ratio chi square = 3.52193 DF = 5 P = .620
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
The final model has generating class
INCCAT*TREAT1*CESDCAT
INCCAT*SEX1
SEX1*TREAT1
The Iterative Proportional Fit algorithm converged at iteration 0.
The maximum difference between observed and fitted marginal totals is .083
and the convergence criterion is .250
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Observed, Expected Frequencies and Residuals.
Factor Code OBS count EXP count Residual Std Resid
INCCAT 0
SEX1 0
TREAT1 0
CESDCAT 0 16.0 13.7 2.28 .62
CESDCAT 1 4.0 6.2 -2.22 -.89
TREAT1 1
CESDCAT 0 23.0 22.2 .82 .17
CESDCAT 1 11.0 11.9 -.88 -.26
SEX1 1
TREAT1 0
CESDCAT 0 48.0 50.3 -2.29 -.32
CESDCAT 1 25.0 22.8 2.21 .46
TREAT1 1
CESDCAT 0 33.0 33.8 -.81 -.14
CESDCAT 1 19.0 18.1 .89 .21
INCCAT 1
SEX1 0
TREAT1 0
CESDCAT 0 16.0 13.8 2.21 .60
CESDCAT 1 5.0 7.3 -2.28 -.84
TREAT1 1
CESDCAT 0 30.0 29.9 .05 .01
CESDCAT 1 6.0 6.0 .01 .00
SEX1 1
TREAT1 0
CESDCAT 0 20.0 22.2 -2.21 -.47
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * *
Observed, Expected Frequencies and Residuals. (Cont.)
Factor Code OBS count EXP count Residual Std Resid
CESDCAT 1 14.0 11.7 2.28 .67
TREAT1 1
CESDCAT 0 20.0 20.1 -.06 -.01
CESDCAT 1 4.0 4.0 -.01 -.01
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Goodness-of-fit test statistics
Likelihood ratio chi square = 3.52193 DF = 5 P = .620
Pearson chi square = 3.37768 DF = 5 P = .642
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Page 430
NOTE: Because a /design subcommand was omitted, SPSS assumes a saturated model.
hiloglinear inccat (0 1) sex1 (0 1) treat1 (0 1) cesdcat (0 1) /print=all.
<some output omitted>
* * * * * * * * H I E R A R C H I C A L L O G L I N E A R * * * * * * * * Tests of PARTIAL associations. Effect Name DF Partial Chisq Prob Iter INCCAT*SEX1*TREAT1 1 .008 .9294 4 INCCAT*SEX1*CESDCAT 1 .009 .9225 3 INCCAT*TREAT1*CESDCAT 1 4.742 .0294 2 SEX1*TREAT1*CESDCAT 1 1.227 .2681 3 INCCAT*SEX1 1 9.906 .0016 3 INCCAT*TREAT1 1 .002 .9653 3 SEX1*TREAT1 1 11.976 .0005 3 INCCAT*CESDCAT 1 1.168 .2799 4 SEX1*CESDCAT 1 2.342 .1259 4 TREAT1*CESDCAT 1 .317 .5733 4 INCCAT 1 14.044 .0002 2 SEX1 1 17.813 .0000 2 TREAT1 1 .014 .9071 2 CESDCAT 1 48.722 .0000 2 - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Page 435
if (sex = 1) sex1= 1. if (sex = 2) sex1 = -1. if (treat =1) treat1 = 1. if (treat = 2) treat1 = -1. if (inccat = 0) inccat1 = -1. if (inccat = 1) inccat1 = 1. compute sextreat = sex1*treat1. compute sexinc = sex1*inccat1. compute treatinc = treat1*inccat1. execute. genlog sex inccat treat with sex1 inccat1 treat1 sexinc sextreat treatinc /model = poisson /print = estim /plot = none /design inccat1 sex1 sexinc sextreat treat1 treatinc.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
GENERAL LOGLINEAR ANALYSIS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Data Information
294 cases are accepted.
0 cases are rejected because of missing data.
294 weighted cases will be used in the analysis.
8 cells are defined.
0 structural zeros are imposed by design.
0 sampling zeros are encountered.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Variable Information
Factor Levels Value
SEX 2
1.00 male
2.00 female
INCCAT 2
.00
1.00
TREAT 2 Has a doctor prescribed or recommended that you take
1.00 yes
2.00 no
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Covariates
SEX1
INCCAT1
TREAT1
SEXINC
SEXTREAT
TREATINC
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Model and Design Information
Model: Poisson
Design: Constant + INCCAT1 + SEX1 + SEXINC + SEXTREAT + TREAT1 + TREATINC
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
GENERAL LOGLINEAR ANALYSIS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Correspondence Between Parameters and Terms of the Design
Parameter Aliased Term
1 Constant
2 INCCAT1
3 SEX1
4 SEXINC
5 SEXTREAT
6 TREAT1
7 TREATINC
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Convergence Information
Maximum number of iterations: 20
Relative difference tolerance: .001
Final relative difference: .0003
Maximum likelihood estimation converged at iteration 2.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Goodness-of-fit Statistics
Chi-Square DF Sig.
Likelihood Ratio .0012 1 .9726
Pearson .0012 1 .9726
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
GENERAL LOGLINEAR ANALYSIS
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Parameter Estimates
Asymptotic 95% CI
Parameter Estimate SE Z-value Lower Upper
1 3.5121 .0636 55.25 3.39 3.64
2 -.1784 .0620 -2.88 -.30 -.06
3 -.2246 .0636 -3.53 -.35 -.10
4 .2056 .0633 3.25 .08 .33
5 -.2194 .0630 -3.48 -.34 -.10
6 -.0481 .0627 -.77 -.17 .07
7 .0005 .0623 8.324E-03 -.12 .12
Page 437
get file 'c:cama3depress.sav'. compute sex1 = sex - 1. compute treat1 = treat - 1. logistic regression var = treat1 with sex1, inccat.