Page 410 Table 16.2
get file 'c:pma5cluster.sav'. descriptives var = ror5 de salesgr5 eps5 npm1 pe payoutr1 /save. list symbol obsno zror5 zde zsalesgr5 znpm1 zpe zpayoutr1.
dia 1.00 .96293 -.00736 .42907 1.28929 -.23733 .15057 dow 2.00 .96293 -.00736 .05048 1.33381 -.44192 -.21655 stf 3.00 .96293 -.55933 -.29025 1.60093 -.44192 -.00701 dd 4.00 .66060 -.92731 -.49217 .48794 -.23733 1.28945 uk 5.00 -.17082 -.55933 -.40383 1.06669 -1.05570 -.66757 psm 6.00 -.24640 -.37534 -.59312 -.22438 -.85111 .82860 gra 7.00 -.20861 -.37534 -.83289 .22082 -.03273 -.23085 hpc 8.00 -.05744 -.74332 -.68146 .08726 -.23733 .59662 mtc 9.00 -.35978 -.55933 -.41645 -.35794 .17186 1.33015 acy 10.00 -.20861 -.55933 -.59312 .13178 -.23733 .66787 cz 11.00 -.96444 -.55933 -.75718 -.40246 -.64652 .65447 ald 12.00 -1.19119 -.19135 -.17667 .35438 -.64652 -1.08741 rom 13.00 -1.00224 -.55933 -.73194 .57698 -.64652 -.73905 rei 14.00 -1.49353 -.55933 .23977 -1.33737 -.03273 -.18999 hum 15.00 -.47315 3.67244 2.90251 .66601 2.21779 -.13474 hca 16.00 -.58653 .36062 1.38816 1.06669 2.42239 -1.97891 nme 17.00 -.77549 .91259 2.76370 .26534 1.80861 -.84038 ami 18.00 -.54874 .72860 .66884 .75505 1.80861 -.83807 ahs 19.00 .92514 -.74332 -.10096 .62149 .78564 -.96513 lks 20.00 1.79435 -.00736 -.18929 -1.24833 -.44192 1.53644 win 21.00 3.00368 -.92731 -.22715 -1.20381 -.23733 1.37083 sgl 22.00 -.20861 1.64855 -.90861 -1.47093 -.85111 -1.70857 slc 23.00 -.20861 .72860 .13882 -1.20381 -.44192 -.69548 kr 24.00 -.09524 -.37534 -.53002 -1.51545 -.85111 .37012 sa 25.00 -.47315 .54461 -.65622 -1.55997 -.64652 1.50460 Number of cases read: 25 Number of cases listed: 25
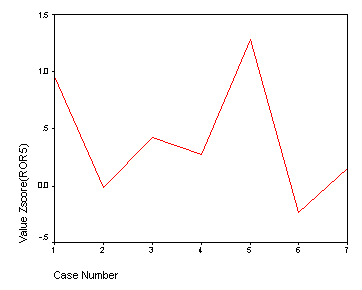
Page 411 Figure 16.3
varstocases /make xaxis from zror5 zde zsalesgr5 zeps5 znpm1 zpe zpayoutr1. use 1 thru 7. graph /line(simple)=value(xaxis).
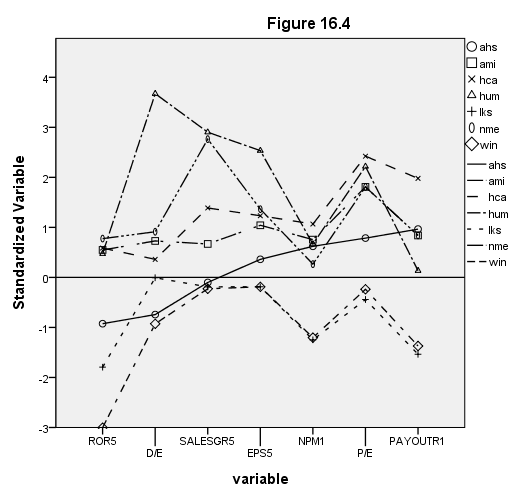
Page 412 Figure 16.4
NOTE: The execute command after the filter command is necessary for the filter to work correctly.
get file 'c:pma5cluster.sav'. descriptives var = ror5 de salesgr5 eps5 npm1 pe payoutr1 /save. compute zror51 = -zror5. compute zpay1 = -zpayoutr1. varstocases /make yaxis from zror51 zde zsalesgr zeps5 znpm1 zpe zpay1 /index = variable. compute filter = (obsno ge 15 and obsno le 21). exe. filter by filter.
value labels variable 1 "ROR5" 2 "D/E" 3 "SALESGR5" 4 "EPS5" 5 "NPM1" 6 "P/E" 7 "PAYOUTR1". formats yaxis (f2.0) . GGRAPH /GRAPHDATASET NAME="GraphDataset" VARIABLES= yaxis variable symbol /GRAPHSPEC SOURCE=INLINE . BEGIN GPL SOURCE: s=userSource( id( "GraphDataset" ) ) DATA: yaxis=col( source(s), name( "yaxis" ) ) DATA: variable=col( source(s), name( "variable" ), unit.category() ) DATA: symbol=col( source(s), name( "symbol" ), unit.category() ) GUIDE: text.title( label( "Figure 16.4" ) ) GUIDE: axis( dim( 1 ), label( "variable" ) ) GUIDE: axis( dim( 2 ), label( "Standardized Variable" ) ) GUIDE: form.line(position(*, 0)) SCALE: linear( dim( 2 ), min(-3), max(4) ) ELEMENT: point( position(summary.mean( variable * yaxis)) ), shape(symbol)) ELEMENT: line( position(summary.mean( variable * yaxis)) ), shape(symbol)) END GPL. filter off.
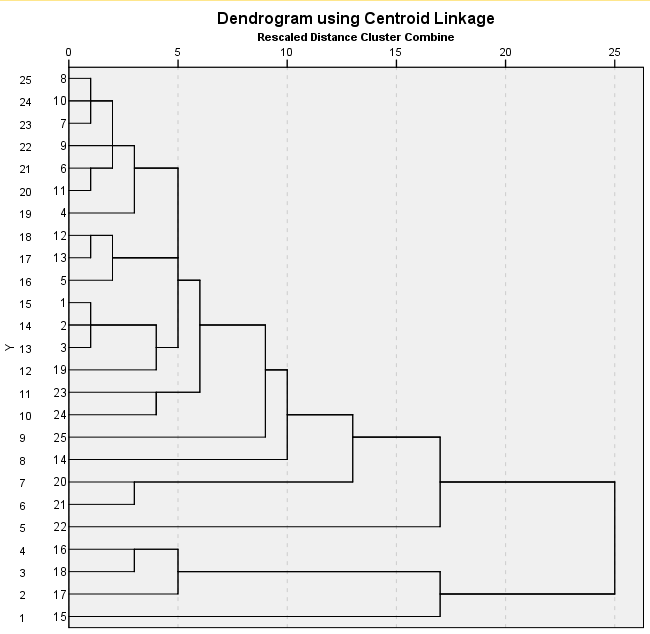
Page 421 Figure 16.9. This does not perfectly reproduce the book.
get file 'c:pma5cluster.sav'. descriptives var = ror5 de salesgr5 eps5 npm1 pe payoutr1 /save. cluster zror5 zde zsalesgr5 zeps5 znpm1 zpe zpayoutr1 /measure = seuclid /method = centroid /plot = dendrogram.
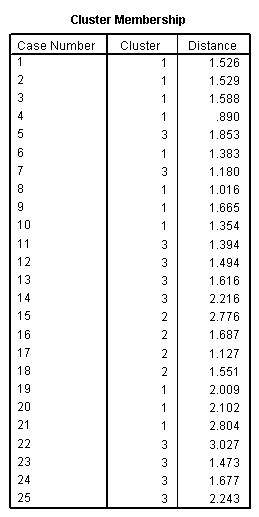
Page 423 Table 16.4
quick cluster zror5 zde zsalesgr5 zeps5 znpm1 zpe zpayoutr1 /criteria = cluster (3) /method = kmeans(noupdate) /print initial cluster distan.
<some output omitted>
Page 424 Figure 16.10
NOTE: This graph has been skipped for now.
