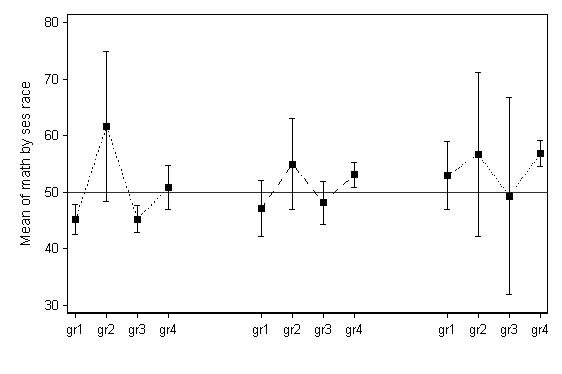
The code below shows how to plot the means and confidence interval bars for groups defined by two categorical variables. This type of plot appeared in an article by Baker, et al, in The American Journal of Clinical Nutrition, "High prepregnant body mass index is associated with early termination of full and any breastfeeding in Danish women".
We start by generating the "bare bones" version of such a graph.
use https://stats.idre.ucla.edu/stat/stata/notes/hsb2, clear preserve local varname math local group ses race collapse (mean) y = `varname' (semean) se_y = `varname', by(`group') sort `group' gen x = _n replace x = _n + 2 if _n >= 5 replace x = _n + 4 if _n >= 9 gen yu = y + 1.96*se_y gen yl = y - 1.96*se_y label define x 1 "gr1" 2 "gr2" 3 "gr3" 4 "gr4" /// 7 "gr1" 8 "gr2" 9 "gr3" 10 "gr4" /// 13 "gr1" 14 "gr2" 15 "gr3" 16 "gr4" label value x x twoway (scatter y x if x<=4, msymbol(S) ) /// (rcap yu yl x if x<=4) (line y x if x<=4) /// (scatter y x if x>=7 & x<=10, msymbol(S) ) /// (rcap yu yl x if x>=7 & x<=10) (line y x if x>=7 & x<=10) /// (scatter y x if x>=13, msymbol(S) ) /// (rcap yu yl x if x>=13) (line y x if x>=13 ), /// xlabel(1(1)4 7(1)10 13(1)16, valuelabel) xtitle(" ") /// ytitle("Mean of `varname' by `group'") yline(50) legend(off) scheme(lean1)
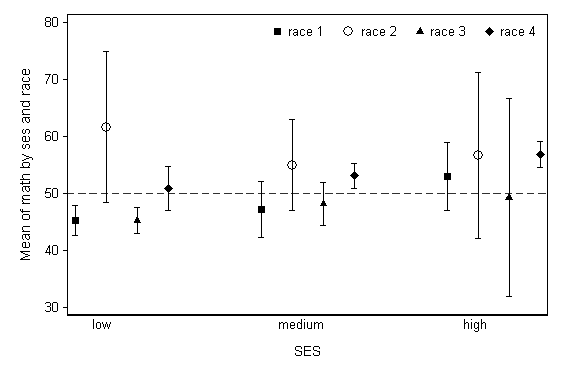
While this graph is of the general desired form, it not clear from looking at it if there are three levels of SES and four levels of race, or vice versa. Ideally, different markers would be used for each of the four levels seen in one group of points, the labeling would be more explicit, and the dotted lines attaching the means would be removed. The code below is more complex, but produces this ideal graph.
restore local varname math local group1 ses local group2 race collapse (mean) y = `varname' (semean) se_y = `varname', by(`group1' `group2') sort `group1' `group2' gen x = _n replace x = _n + 2 if _n >= 5 replace x = _n + 4 if _n >= 9 gen yu = y + 1.96*se_y gen yl = y - 1.96*se_y separate y, by(`group2') separate yu, by(`group2') separate yl, by(`group2') label define x 1 "gr1" 2 "gr2" 3 "gr3" 4 "gr4" /// 7 "gr1" 8 "gr2" 9 "gr3" 10 "gr4" /// 13 "gr1" 14 "gr2" 15 "gr3" 16 "gr4" label value x x local varname math local group1 ses local group2 race twoway (scatter y1 x , msymbol(S) msize(medium)) /// (rcap yu1 yl1 x) /// (scatter y2 x , msymbol(Oh) msize(large)) /// (rcap yu2 yl2 x ) /// (scatter y3 x , msymbol(T) msize(medium)) /// (rcap yu3 yl3 x ) /// (scatter y4 x , msymbol(D) msize(medium)) /// (rcap yu4 yl4 x ), scheme(lean1) /// xlabel(none) xtitle(" ") b2("SES") /// ytitle(Mean of `varname' by `group1' and `group2') yline(50, lpattern(dash)) /// text(27 1.5 "low", place(e)) /// text(27 7.5 "medium", place(e)) /// text(27 13.5 "high", place(e)) /// legend(order(1 "race 1" 3 "race 2" 5 "race 3" 7 "race 4") position(1) row(1) ring(0)) /// name(g3, replace)
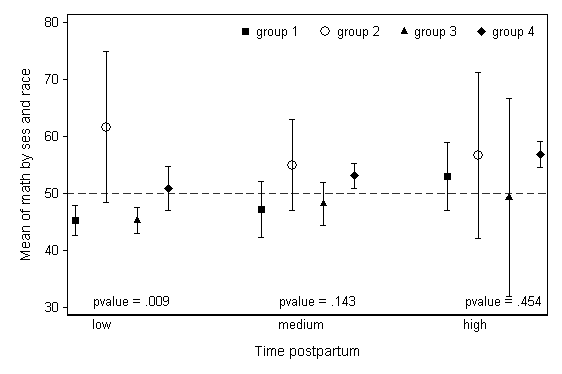
We may wish to add additional text to this plot, like p-values from ANOVA tests on race conducted within SES levels. The code below starts from the original dataset to generate these values, saves them as macro variables, and then puts them into the plot using the text option (similar to the placement of SES levels in the above plot).
use https://stats.idre.ucla.edu/stat/stata/notes/hsb2, clear anova math race if ses==1 test race local pv1 = round(Ftail(r(df), r(df_r),r(F)), .001) anova math race if ses==2 test race local pv2 = round(Ftail(r(df), r(df_r),r(F)), .001) anova math race if ses==3 test race local pv3 = round(Ftail(r(df), r(df_r),r(F)), .001) local varname math local group1 ses local group2 race collapse (mean) y = `varname' (semean) se_y = `varname', by(`group1' `group2') sort `group1' `group2' gen x = _n replace x = _n + 2 if _n >= 5 replace x = _n + 4 if _n >= 9 gen yu = y + 1.96*se_y gen yl = y - 1.96*se_y separate y, by(`group2') separate yu, by(`group2') separate yl, by(`group2') * no connecting lines, another version local varname math local group1 ses local group2 race di `pv1' twoway (scatter y1 x , msymbol(S) msize(medium)) /// (rcap yu1 yl1 x) /// (scatter y2 x , msymbol(Oh) msize(large)) /// (rcap yu2 yl2 x ) /// (scatter y3 x , msymbol(T) msize(medium)) /// (rcap yu3 yl3 x ) /// (scatter y4 x , msymbol(D) msize(medium)) /// (rcap yu4 yl4 x ), scheme(lean1) /// xlabel(none) xtitle(" ") b2("Time postpartum") /// ytitle(Mean of `varname' by `group1' and `group2') yline(50, lpattern(dash)) /// text(31 1.5 "p = `pv1'", place(e)) /// text(31 7.5 "p = `pv2'", place(e)) /// text(31 13.5 "p = `pv3'", place(e)) /// text(27 1.5 "low", place(e)) /// text(27 7.5 "medium", place(e)) /// text(27 13.5 "high", place(e)) /// legend(order(1 "group 1" 3 "group 2" 5 "group 3" 7 "group 4") position(1) row(1) ring(0))