Table 5.1, page 150.
use uis.dta, clear
gen ndrgfp1 = ((ndrugtx+1)/10)^(-1)
gen ndrgfp2 = ndrgfp1*log((ndrugtx+1)/10)
gen agendrgfp1 = age*ndrgfp1
gen racesite = race*site
quietly logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
* Stata 8 code.
lfit, table group(10)
* Stata 9 code and output.
estat gof, table group(10)
Logistic model for dfree, goodness-of-fit test
(Table collapsed on quantiles of estimated probabilities)
+--------------------------------------------------------+
| Group | Prob | Obs_1 | Exp_1 | Obs_0 | Exp_0 | Total |
|-------+--------+-------+-------+-------+-------+-------|
| 1 | 0.0939 | 4 | 4.1 | 54 | 53.9 | 58 |
| 2 | 0.1261 | 5 | 6.2 | 52 | 50.8 | 57 |
| 3 | 0.1631 | 8 | 8.5 | 50 | 49.5 | 58 |
| 4 | 0.2036 | 11 | 10.4 | 46 | 46.6 | 57 |
| 5 | 0.2335 | 16 | 12.7 | 42 | 45.3 | 58 |
|-------+--------+-------+-------+-------+-------+-------|
| 6 | 0.2788 | 11 | 14.5 | 46 | 42.5 | 57 |
| 7 | 0.3240 | 18 | 17.5 | 40 | 40.5 | 58 |
| 8 | 0.3764 | 24 | 19.8 | 33 | 37.2 | 57 |
| 9 | 0.4590 | 23 | 23.9 | 35 | 34.1 | 58 |
| 10 | 0.7283 | 27 | 29.3 | 30 | 27.7 | 57 |
+--------------------------------------------------------+
number of observations = 575
number of groups = 10
Hosmer-Lemeshow chi2(8) = 4.39
Prob > chi2 = 0.8199
Table 5.2, page 157.
* Stata 8 code.
lstat
* Stata 9 code and output.
estat classification
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 16 11 | 27
- | 131 417 | 548
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .5
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 10.88%
Specificity Pr( -|~D) 97.43%
Positive predictive value Pr( D| +) 59.26%
Negative predictive value Pr(~D| -) 76.09%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 2.57%
False - rate for true D Pr( -| D) 89.12%
False + rate for classified + Pr(~D| +) 40.74%
False - rate for classified - Pr( D| -) 23.91%
--------------------------------------------------
Correctly classified 75.30%
--------------------------------------------------
Table 5.3, page 159.
NOTE: We could not recreate this table.
Table 5.4, page 160.
NOTE: We could not recreate this table.
Table 5.5, page 161.
quietly logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
* Stata 8 code.
lstat, cutoff(.6)
* Stata 9 code and output.
estat classification, cutoff(.6)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 5 0 | 5
- | 142 428 | 570
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .6
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 3.40%
Specificity Pr( -|~D) 100.00%
Positive predictive value Pr( D| +) 100.00%
Negative predictive value Pr(~D| -) 75.09%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 0.00%
False - rate for true D Pr( -| D) 96.60%
False + rate for classified + Pr(~D| +) 0.00%
False - rate for classified - Pr( D| -) 24.91%
--------------------------------------------------
Correctly classified 75.30%
--------------------------------------------------
Table 5.6, page 161.
quietly logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
* Stata 8 code.
lstat, cutoff(.05)
* Stata 9 code and output.
estat classification, cutoff(.05)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 146 417 | 563
- | 1 11 | 12
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .05
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 99.32%
Specificity Pr( -|~D) 2.57%
Positive predictive value Pr( D| +) 25.93%
Negative predictive value Pr(~D| -) 91.67%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 97.43%
False - rate for true D Pr( -| D) 0.68%
False + rate for classified + Pr(~D| +) 74.07%
False - rate for classified - Pr( D| -) 8.33%
--------------------------------------------------
Correctly classified 27.30%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.1)
* Stata 9 code and output.
estat classification, cutoff(.1)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 141 363 | 504
- | 6 65 | 71
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .1
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 95.92%
Specificity Pr( -|~D) 15.19%
Positive predictive value Pr( D| +) 27.98%
Negative predictive value Pr(~D| -) 91.55%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 84.81%
False - rate for true D Pr( -| D) 4.08%
False + rate for classified + Pr(~D| +) 72.02%
False - rate for classified - Pr( D| -) 8.45%
--------------------------------------------------
Correctly classified 35.83%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.15)
* Stata 9 code and output.
estat classification, cutoff(.15)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 133 292 | 425
- | 14 136 | 150
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .15
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 90.48%
Specificity Pr( -|~D) 31.78%
Positive predictive value Pr( D| +) 31.29%
Negative predictive value Pr(~D| -) 90.67%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 68.22%
False - rate for true D Pr( -| D) 9.52%
False + rate for classified + Pr(~D| +) 68.71%
False - rate for classified - Pr( D| -) 9.33%
--------------------------------------------------
Correctly classified 46.78%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.2)
* Stata 9 code and output.
estat classification, cutoff(.2)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 120 230 | 350
- | 27 198 | 225
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .2
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 81.63%
Specificity Pr( -|~D) 46.26%
Positive predictive value Pr( D| +) 34.29%
Negative predictive value Pr(~D| -) 88.00%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 53.74%
False - rate for true D Pr( -| D) 18.37%
False + rate for classified + Pr(~D| +) 65.71%
False - rate for classified - Pr( D| -) 12.00%
--------------------------------------------------
Correctly classified 55.30%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.25)
* Stata 9 code and output.
estat classification, cutoff(.25)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 97 166 | 263
- | 50 262 | 312
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .25
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 65.99%
Specificity Pr( -|~D) 61.21%
Positive predictive value Pr( D| +) 36.88%
Negative predictive value Pr(~D| -) 83.97%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 38.79%
False - rate for true D Pr( -| D) 34.01%
False + rate for classified + Pr(~D| +) 63.12%
False - rate for classified - Pr( D| -) 16.03%
--------------------------------------------------
Correctly classified 62.43%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.3)
* Stata 9 code and output.
estat classification, cutoff(.3)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 84 119 | 203
- | 63 309 | 372
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .3
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 57.14%
Specificity Pr( -|~D) 72.20%
Positive predictive value Pr( D| +) 41.38%
Negative predictive value Pr(~D| -) 83.06%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 27.80%
False - rate for true D Pr( -| D) 42.86%
False + rate for classified + Pr(~D| +) 58.62%
False - rate for classified - Pr( D| -) 16.94%
--------------------------------------------------
Correctly classified 68.35%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.35)
* Stata 9 code and output.
estat classification, cutoff(.35)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 59 77 | 136
- | 88 351 | 439
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .35
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 40.14%
Specificity Pr( -|~D) 82.01%
Positive predictive value Pr( D| +) 43.38%
Negative predictive value Pr(~D| -) 79.95%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 17.99%
False - rate for true D Pr( -| D) 59.86%
False + rate for classified + Pr(~D| +) 56.62%
False - rate for classified - Pr( D| -) 20.05%
--------------------------------------------------
Correctly classified 71.30%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.4)
* Stata 9 code and output.
estat classification, cutoff(.4)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 43 54 | 97
- | 104 374 | 478
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .4
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 29.25%
Specificity Pr( -|~D) 87.38%
Positive predictive value Pr( D| +) 44.33%
Negative predictive value Pr(~D| -) 78.24%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 12.62%
False - rate for true D Pr( -| D) 70.75%
False + rate for classified + Pr(~D| +) 55.67%
False - rate for classified - Pr( D| -) 21.76%
--------------------------------------------------
Correctly classified 72.52%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.45)
* Stata 9 code and output.
estat classification, cutoff(.45)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 27 34 | 61
- | 120 394 | 514
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .45
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 18.37%
Specificity Pr( -|~D) 92.06%
Positive predictive value Pr( D| +) 44.26%
Negative predictive value Pr(~D| -) 76.65%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 7.94%
False - rate for true D Pr( -| D) 81.63%
False + rate for classified + Pr(~D| +) 55.74%
False - rate for classified - Pr( D| -) 23.35%
--------------------------------------------------
Correctly classified 73.22%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.5)
* Stata 9 code and output.
estat classification, cutoff(.5)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 16 11 | 27
- | 131 417 | 548
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .5
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 10.88%
Specificity Pr( -|~D) 97.43%
Positive predictive value Pr( D| +) 59.26%
Negative predictive value Pr(~D| -) 76.09%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 2.57%
False - rate for true D Pr( -| D) 89.12%
False + rate for classified + Pr(~D| +) 40.74%
False - rate for classified - Pr( D| -) 23.91%
--------------------------------------------------
Correctly classified 75.30%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.55)
* Stata 9 code and output.
estat classification, cutoff(.55)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 8 3 | 11
- | 139 425 | 564
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .55
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 5.44%
Specificity Pr( -|~D) 99.30%
Positive predictive value Pr( D| +) 72.73%
Negative predictive value Pr(~D| -) 75.35%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 0.70%
False - rate for true D Pr( -| D) 94.56%
False + rate for classified + Pr(~D| +) 27.27%
False - rate for classified - Pr( D| -) 24.65%
--------------------------------------------------
Correctly classified 75.30%
--------------------------------------------------
* Stata 8 code.
lstat, cutoff(.6)
* Stata 9 code and output.
estat classification, cutoff(.6)
Logistic model for dfree
-------- True --------
Classified | D ~D | Total
-----------+--------------------------+-----------
+ | 5 0 | 5
- | 142 428 | 570
-----------+--------------------------+-----------
Total | 147 428 | 575
Classified + if predicted Pr(D) >= .6
True D defined as dfree ~= 0
--------------------------------------------------
Sensitivity Pr( +| D) 3.40%
Specificity Pr( -|~D) 100.00%
Positive predictive value Pr( D| +) 100.00%
Negative predictive value Pr(~D| -) 75.09%
--------------------------------------------------
False + rate for true ~D Pr( +|~D) 0.00%
False - rate for true D Pr( -| D) 96.60%
False + rate for classified + Pr(~D| +) 0.00%
False - rate for classified - Pr( D| -) 24.91%
--------------------------------------------------
Correctly classified 75.30%
--------------------------------------------------
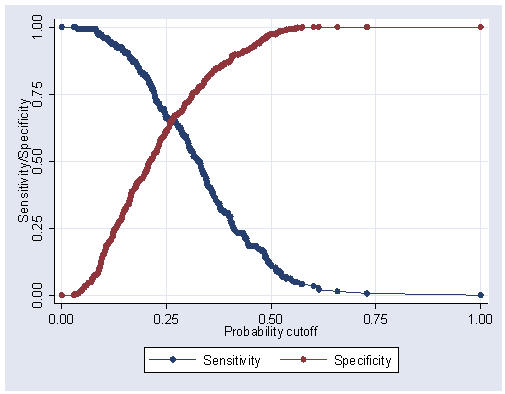
Figure 5.1, page 162.
lsens
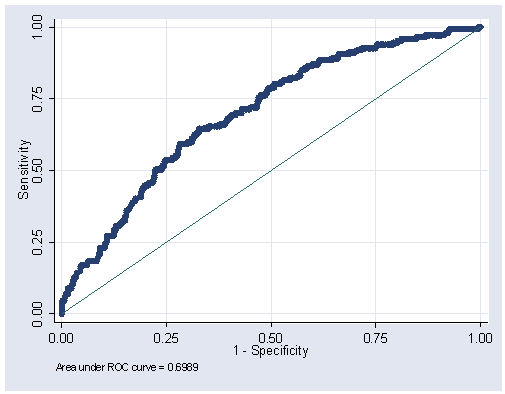
Figure 5.2, page 163.
lroc
Logistic model for dfree number of observations = 575 area under ROC curve = 0.6989
Figure 5.3, page 171.
NOTE: We cannot recreate this figure because we do not have the hypothetical data that were used.
Figure 5.4, page 172.
NOTE: We cannot recreate this figure because we do not have the hypothetical data that were used.
Figure 5.5, page 177.
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
Iteration 0: log likelihood = -326.86446
Iteration 1: log likelihood = -300.06724
Iteration 2: log likelihood = -298.98837
Iteration 3: log likelihood = -298.98146
Iteration 4: log likelihood = -298.98146
Logit estimates Number of obs = 575
LR chi2(10) = 55.77
Prob > chi2 = 0.0000
Log likelihood = -298.98146 Pseudo R2 = 0.0853
------------------------------------------------------------------------------
dfree | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .1166385 .0288749 4.04 0.000 .0600446 .1732323
ndrgfp1 | 1.669035 .407152 4.10 0.000 .871032 2.467038
ndrgfp2 | .4336886 .1169052 3.71 0.000 .2045586 .6628185
ivhx2 | -.6346307 .2987192 -2.12 0.034 -1.220109 -.0491518
ivhx3 | -.7049475 .2615805 -2.69 0.007 -1.217636 -.1922591
race | .6841068 .2641355 2.59 0.010 .1664107 1.201803
treat | .4349255 .2037596 2.13 0.033 .035564 .834287
site | .516201 .2548881 2.03 0.043 .0166295 1.015773
agendrgfp1 | -.0152697 .0060268 -2.53 0.011 -.0270819 -.0034575
racesite | -1.429457 .5297806 -2.70 0.007 -2.467808 -.3911062
_cons | -6.843864 1.219316 -5.61 0.000 -9.23368 -4.454048
------------------------------------------------------------------------------
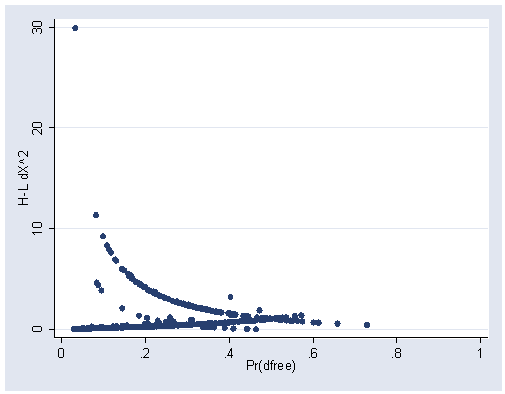
predict p
(option p assumed; Pr(dfree))
predict dx, dx2
graph twoway scatter dx p, xlabel(0(.2)1) ylabel(0(10)30)
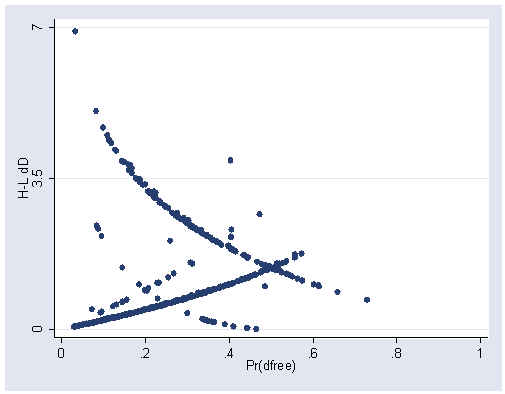
Figure 5.6, page 178.
predict dd, dd graph twoway scatter dd p, xlabel(0(.2)1) ylabel(0 3.5 7)
Figure 5.7, page 179.
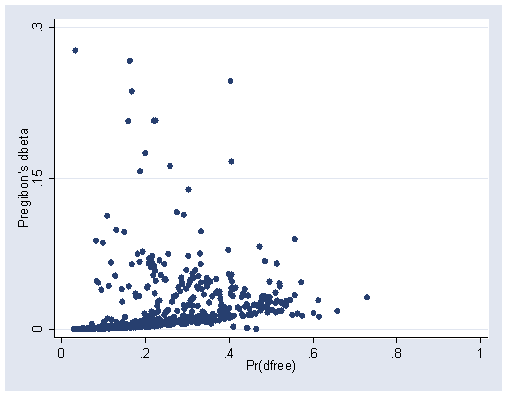
predict db, db graph twoway scatter db p, xlabel(0(.2)1) ylabel(0 .15 .3)
Figure 5.8, page 180.
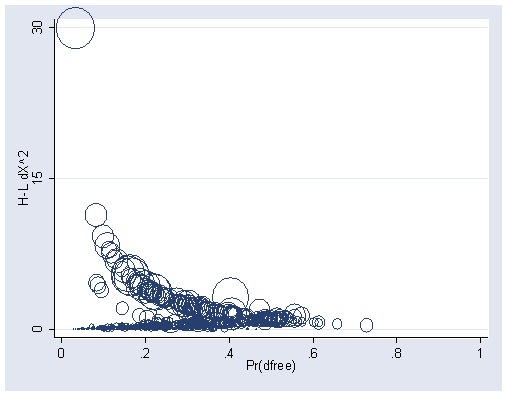
graph twoway scatter dx p [weight=db], xlabel(0(.2)1) ylabel(0 15 30) msymbol(oh)
Table 5.8, page 182.
predict h, h predict n, n list age ndrugtx ivhx2 race treat site dfree n p db dx dd h if n==31 | n==477 | n==105 | n==468
Covariate pattern 105
Observation 84
age 26 ndrugtx 0 ivhx2 0
race 1 treat 0 site 0
dfree 1 n 105 p .4030007
db .2462623 dx 3.191391 dd 3.915781
h .0716367
Covariate pattern 468
Observation 351
age 40 ndrugtx 0 ivhx2 0
race 1 treat 0 site 0
dfree 1 n 468 p .1675982
db .2363966 dx 5.192755 dd 3.735002
h .0435421
Covariate pattern 477
Observation 367
age 41 ndrugtx 0 ivhx2 0
race 1 treat 0 site 0
dfree 1 n 477 p .1626278
db .266626 dx 5.403098 dd 3.811839
h .0470263
Covariate pattern 31
Observation 519
age 24 ndrugtx 20 ivhx2 1
race 0 treat 0 site 1
dfree 1 n 31 p .0326259
db .2768316 dx 29.92479 dd 6.908623
h .0091661
Covariate pattern 105
Observation 548
age 26 ndrugtx 0 ivhx2 0
race 1 treat 0 site 0
dfree 1 n 105 p .4030007
db .2462623 dx 3.191391 dd 3.915781
h .0716367
NOTE: There are five cases listed because covariate pattern 105 has two cases; only one of them is listed in the text.
Table 5.9, page 183.
NOTE: The goodness-of-fit values at the bottom of the table are not percent change; they are the actual values obtained from the goodness-of-fit tests.
NOTE: Use the ldev command to get the D value. Use the lfit, group(10) command to get the chi-square value. Use the lfit command to get the C-hat value. All data (note that this column contains the coefficients from the logit):
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
Iteration 0: log likelihood = -326.86446
Iteration 1: log likelihood = -300.06724
Iteration 2: log likelihood = -298.98837
Iteration 3: log likelihood = -298.98146
Iteration 4: log likelihood = -298.98146
Logit estimates Number of obs = 575
LR chi2(10) = 55.77
Prob > chi2 = 0.0000
Log likelihood = -298.98146 Pseudo R2 = 0.0853
------------------------------------------------------------------------------
dfree | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .1166385 .0288749 4.04 0.000 .0600446 .1732323
ndrgfp1 | 1.669035 .407152 4.10 0.000 .871032 2.467038
ndrgfp2 | .4336886 .1169052 3.71 0.000 .2045586 .6628185
ivhx2 | -.6346307 .2987192 -2.12 0.034 -1.220109 -.0491518
ivhx3 | -.7049475 .2615805 -2.69 0.007 -1.217636 -.1922591
race | .6841068 .2641355 2.59 0.010 .1664107 1.201803
treat | .4349255 .2037596 2.13 0.033 .035564 .834287
site | .516201 .2548881 2.03 0.043 .0166295 1.015773
agendrgfp1 | -.0152697 .0060268 -2.53 0.011 -.0270819 -.0034575
racesite | -1.429457 .5297806 -2.70 0.007 -2.467808 -.3911062
_cons | -6.843864 1.219316 -5.61 0.000 -9.23368 -4.454048
------------------------------------------------------------------------------
* Stata 8 code.
lfit
* Stata 9 code and output.
estat gof
Logistic model for dfree, goodness-of-fit test
number of observations = 575
number of covariate patterns = 521
Pearson chi2(510) = 511.78
Prob > chi2 = 0.4695
The ldev command can be installed from the ATS web site by typing search ldev (see How can I use the search command to search for programs and get additional help? for more information about using search).
ldev
Logistic model deviance goodness-of-fit test
number of observations = 575
number of covariate patterns = 521
deviance goodness-of-fit = 530.74
degrees of freedom = 510
Prob > chi2 = 0.2541
* Stata 8 code.
lfit, group(10) table
* Stata 9 code and output.
estat gof, group(10) table
Logistic model for dfree, goodness-of-fit test
(Table collapsed on quantiles of estimated probabilities)
_Group _Prob _Obs_1 _Exp_1 _Obs_0 _Exp_0 _Total
1 0.0939 4 4.1 54 53.9 58
2 0.1261 5 6.2 52 50.8 57
3 0.1631 8 8.5 50 49.5 58
4 0.2036 11 10.4 46 46.6 57
5 0.2335 16 12.7 42 45.3 58
6 0.2788 11 14.5 46 42.5 57
7 0.3240 18 17.5 40 40.5 58
8 0.3764 24 19.8 33 37.2 57
9 0.4590 23 23.9 35 34.1 58
10 0.7283 27 29.3 30 27.7 57
number of observations = 575
number of groups = 10
Hosmer-Lemeshow chi2(8) = 4.39
Prob > chi2 = 0.8199
Deleting covariate pattern 31 (note that this column contains percent change between the coefficients from the two logits).
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite if n!=31
Iteration 0: log likelihood = -325.49798
Iteration 1: log likelihood = -296.7082
Iteration 2: log likelihood = -295.42918
Iteration 3: log likelihood = -295.41908
Iteration 4: log likelihood = -295.41908
Logit estimates Number of obs = 574
LR chi2(10) = 60.16
Prob > chi2 = 0.0000
Log likelihood = -295.41908 Pseudo R2 = 0.0924
------------------------------------------------------------------------------
dfree | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .1269757 .0294627 4.31 0.000 .0692298 .1847216
ndrgfp1 | 1.829975 .4173235 4.39 0.000 1.012036 2.647914
ndrgfp2 | .4746444 .1190842 3.99 0.000 .2412436 .7080452
ivhx2 | -.6904163 .3028271 -2.28 0.023 -1.283947 -.096886
ivhx3 | -.708751 .2629679 -2.70 0.007 -1.224159 -.1933435
race | .6927178 .2656414 2.61 0.009 .1720703 1.213365
treat | .4574146 .2052736 2.23 0.026 .0550858 .8597434
site | .4873447 .257198 1.89 0.058 -.0167541 .9914435
agendrgfp1 | -.0167797 .0061338 -2.74 0.006 -.0288017 -.0047577
racesite | -1.422103 .5322418 -2.67 0.008 -2.465278 -.3789279
_cons | -7.372811 1.253245 -5.88 0.000 -9.829126 -4.916496
------------------------------------------------------------------------------
* Stata 8 code.
lfit
* Stata 9 code and output.
estat gof
Logistic model for dfree, goodness-of-fit test
number of observations = 574
number of covariate patterns = 520
Pearson chi2(509) = 489.94
Prob > chi2 = 0.7204
ldev
(1 missing value generated)
(1 missing value generated)
(1 missing value generated)
Logistic model deviance goodness-of-fit test
number of observations = 574
number of covariate patterns = 520
deviance goodness-of-fit = 523.62
degrees of freedom = 509
Prob > chi2 = 0.3175
* Stata 8 code.
lfit, group(10) table
* Stata 9 code and output.
estat gof, group(10) table
Logistic model for dfree, goodness-of-fit test
(Table collapsed on quantiles of estimated probabilities)
_Group _Prob _Obs_1 _Exp_1 _Obs_0 _Exp_0 _Total
1 0.0872 3 3.8 55 54.2 58
2 0.1217 5 5.9 52 51.1 57
3 0.1578 7 8.2 51 49.8 58
4 0.1999 14 10.3 44 47.7 58
5 0.2322 14 12.1 42 43.9 56
6 0.2725 11 14.6 47 43.4 58
7 0.3247 21 17.2 36 39.8 57
8 0.3775 21 20.3 37 37.7 58
9 0.4602 23 23.8 34 33.2 57
10 0.7480 27 29.9 30 27.1 57
number of observations = 574
number of groups = 10
Hosmer-Lemeshow chi2(8) = 5.55
Prob > chi2 = 0.6973
di (.1269757-.1166385)
.0103372
di (.0103372/.1166385)*100
8.8625968
NOTE: We will not show all of the rest of the percent change calculations; they can be obtained in the same way as above.
Deleting covariate pattern 477.
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite if n!=477
Iteration 0: log likelihood = -325.49798
Iteration 1: log likelihood = -298.19394
Iteration 2: log likelihood = -297.04285
Iteration 3: log likelihood = -297.03469
Iteration 4: log likelihood = -297.03469
Logit estimates Number of obs = 574
LR chi2(10) = 56.93
Prob > chi2 = 0.0000
Log likelihood = -297.03469 Pseudo R2 = 0.0874
------------------------------------------------------------------------------
dfree | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .1228473 .0293551 4.18 0.000 .0653123 .1803822
ndrgfp1 | 1.77494 .4183485 4.24 0.000 .954992 2.594888
ndrgfp2 | .45136 .1183443 3.81 0.000 .2194095 .6833106
ivhx2 | -.6374841 .2990411 -2.13 0.033 -1.223594 -.0513744
ivhx3 | -.7445476 .2636286 -2.82 0.005 -1.26125 -.2278452
race | .6441888 .2660551 2.42 0.015 .1227304 1.165647
treat | .4503906 .2045379 2.20 0.028 .0495036 .8512776
site | .5162495 .2553121 2.02 0.043 .015847 1.016652
agendrgfp1 | -.0175048 .0062929 -2.78 0.005 -.0298386 -.005171
racesite | -1.377475 .5319126 -2.59 0.010 -2.420005 -.3349456
_cons | -7.070646 1.240009 -5.70 0.000 -9.501018 -4.640273
------------------------------------------------------------------------------
* Stata 8 code.
lfit
* Stata 9 code and output.
estat gof
Logistic model for dfree, goodness-of-fit test
number of observations = 574
number of covariate patterns = 520
Pearson chi2(509) = 511.57
Prob > chi2 = 0.4597
ldev
(1 missing value generated)
(1 missing value generated)
(1 missing value generated)
Logistic model deviance goodness-of-fit test
number of observations = 574
number of covariate patterns = 520
deviance goodness-of-fit = 526.85
degrees of freedom = 509
Prob > chi2 = 0.2830
* Stata 8 code.
lfit, group(10) table
* Stata 9 code and output.
estat gof, group(10) table
Logistic model for dfree, goodness-of-fit test
(Table collapsed on quantiles of estimated probabilities)
_Group _Prob _Obs_1 _Exp_1 _Obs_0 _Exp_0 _Total
1 0.0895 4 3.9 54 54.1 58
2 0.1232 5 6.1 52 50.9 57
3 0.1625 8 8.4 50 49.6 58
4 0.2008 11 10.3 46 46.7 57
5 0.2337 16 12.4 41 44.6 57
6 0.2756 9 14.8 49 43.2 58
7 0.3244 20 17.1 37 39.9 57
8 0.3736 23 20.3 35 37.7 58
9 0.4559 23 23.5 34 33.5 57
10 0.7428 27 29.3 30 27.7 57
number of observations = 574
number of groups = 10
Hosmer-Lemeshow chi2(8) = 6.36
Prob > chi2 = 0.6074
Deleting covariate pattern 105.
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite if n!=105
Iteration 0: log likelihood = -324.1264
Iteration 1: log likelihood = -298.0818
Iteration 2: log likelihood = -297.0549
Iteration 3: log likelihood = -297.04873
Iteration 4: log likelihood = -297.04873
Logit estimates Number of obs = 573
LR chi2(10) = 54.16
Prob > chi2 = 0.0000
Log likelihood = -297.04873 Pseudo R2 = 0.0835
------------------------------------------------------------------------------
dfree | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .1134016 .0288724 3.93 0.000 .0568126 .1699905
ndrgfp1 | 1.642961 .4065219 4.04 0.000 .8461929 2.43973
ndrgfp2 | .442747 .1170361 3.78 0.000 .2133604 .6721336
ivhx2 | -.6368155 .2992244 -2.13 0.033 -1.223285 -.0503465
ivhx3 | -.7046331 .2620011 -2.69 0.007 -1.218146 -.1911204
race | .6258739 .2671909 2.34 0.019 .1021893 1.149558
treat | .4669661 .2049928 2.28 0.023 .0651876 .8687445
site | .5309903 .2550417 2.08 0.037 .0311177 1.030863
agendrgfp1 | -.0139969 .0060693 -2.31 0.021 -.0258925 -.0021013
racesite | -1.370085 .531162 -2.58 0.010 -2.411144 -.329027
_cons | -6.756569 1.216512 -5.55 0.000 -9.140889 -4.37225
------------------------------------------------------------------------------
* Stata 8 code.
lfit
* Stata 9 code and output.
estat gof
Logistic model for dfree, goodness-of-fit test
number of observations = 573
number of covariate patterns = 520
Pearson chi2(509) = 508.70
Prob > chi2 = 0.4954
ldev
(2 missing values generated)
(2 missing values generated)
(2 missing values generated)
Logistic model deviance goodness-of-fit test
number of observations = 573
number of covariate patterns = 520
deviance goodness-of-fit = 526.88
degrees of freedom = 509
Prob > chi2 = 0.2828
* Stata 8 code.
lfit, group(10) table
* Stata 9 code and output.
estat gof, group(10) table
Logistic model for dfree, goodness-of-fit test
(Table collapsed on quantiles of estimated probabilities)
_Group _Prob _Obs_1 _Exp_1 _Obs_0 _Exp_0 _Total
1 0.0947 4 4.1 54 53.9 58
2 0.1256 6 6.3 51 50.7 57
3 0.1640 8 8.4 49 48.6 57
4 0.2022 9 10.5 49 47.5 58
5 0.2330 17 12.4 40 44.6 57
6 0.2815 10 14.4 47 42.6 57
7 0.3198 20 17.3 38 40.7 58
8 0.3601 23 19.4 34 37.6 57
9 0.4532 22 23.3 35 33.7 57
10 0.7128 26 29.0 31 28.0 57
number of observations = 573
number of groups = 10
Hosmer-Lemeshow chi2(8) = 6.69
Prob > chi2 = 0.5705
Deleting covariate pattern 468.
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite if n!=468
Iteration 0: log likelihood = -325.49798
Iteration 1: log likelihood = -298.23262
Iteration 2: log likelihood = -297.08748
Iteration 3: log likelihood = -297.07943
Iteration 4: log likelihood = -297.07943
Logit estimates Number of obs = 574
LR chi2(10) = 56.84
Prob > chi2 = 0.0000
Log likelihood = -297.07943 Pseudo R2 = 0.0873
------------------------------------------------------------------------------
dfree | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .1222986 .0293131 4.17 0.000 .064846 .1797511
ndrgfp1 | 1.764847 .4173058 4.23 0.000 .9469426 2.582751
ndrgfp2 | .4502113 .1182347 3.81 0.000 .2184756 .681947
ivhx2 | -.6392831 .2990409 -2.14 0.033 -1.225393 -.0531736
ivhx3 | -.7455178 .2636375 -2.83 0.005 -1.262238 -.2287979
race | .6437812 .266046 2.42 0.016 .1223406 1.165222
treat | .4506826 .2045295 2.20 0.028 .0498121 .8515532
site | .5164371 .2552941 2.02 0.043 .0160699 1.016804
agendrgfp1 | -.0172712 .0062674 -2.76 0.006 -.0295551 -.0049874
racesite | -1.37849 .5318175 -2.59 0.010 -2.420833 -.3361467
_cons | -7.048292 1.238013 -5.69 0.000 -9.474753 -4.621832
------------------------------------------------------------------------------
* Stata 8 code.
lfit
* Stata 9 code and output.
estat gof
Logistic model for dfree, goodness-of-fit test
number of observations = 574
number of covariate patterns = 520
Pearson chi2(509) = 511.61
Prob > chi2 = 0.4591
ldev
(1 missing value generated)
(1 missing value generated)
(1 missing value generated)
Logistic model deviance goodness-of-fit test
number of observations = 574
number of covariate patterns = 520
deviance goodness-of-fit = 526.94
degrees of freedom = 509
Prob > chi2 = 0.2821
* Stata 8 code.
lfit, group(10) table
* Stata 9 code and output.
estat gof, group(10) table
Logistic model for dfree, goodness-of-fit test
(Table collapsed on quantiles of estimated probabilities)
_Group _Prob _Obs_1 _Exp_1 _Obs_0 _Exp_0 _Total
1 0.0899 4 4.0 54 54.0 58
2 0.1232 5 6.1 52 50.9 57
3 0.1623 8 8.4 50 49.6 58
4 0.2011 11 10.3 46 46.7 57
5 0.2332 16 12.6 42 45.4 58
6 0.2761 9 14.8 49 43.2 58
7 0.3230 20 16.8 36 39.2 56
8 0.3740 23 20.3 35 37.7 58
9 0.4565 23 23.5 34 33.5 57
10 0.7414 27 29.3 30 27.7 57
number of observations = 574
number of groups = 10
Hosmer-Lemeshow chi2(8) = 6.36
Prob > chi2 = 0.6065
Deleting all four covariate patterns.
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site ///
agendrgfp1 racesite if n!=31 & n!=477 & n!=105 & n!=468
Iteration 0: log likelihood = -319.98056
Iteration 1: log likelihood = -290.60629
Iteration 2: log likelihood = -289.18
Iteration 3: log likelihood = -289.16639
Iteration 4: log likelihood = -289.16638
Logit estimates Number of obs = 570
LR chi2(10) = 61.63
Prob > chi2 = 0.0000
Log likelihood = -289.16638 Pseudo R2 = 0.0963
------------------------------------------------------------------------------
dfree | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .1375773 .0305755 4.50 0.000 .0776504 .1975042
ndrgfp1 | 2.042452 .4429761 4.61 0.000 1.174235 2.910669
ndrgfp2 | .5253043 .1228263 4.28 0.000 .2845692 .7660395
ivhx2 | -.7016545 .3043009 -2.31 0.021 -1.298073 -.1052357
ivhx3 | -.796238 .2678256 -2.97 0.003 -1.321167 -.2713095
race | .5453942 .2730249 2.00 0.046 .0102753 1.080513
treat | .5253459 .2084363 2.52 0.012 .1168182 .9338736
site | .5042472 .2584389 1.95 0.051 -.0022837 1.010778
agendrgfp1 | -.0204074 .0067651 -3.02 0.003 -.0336668 -.007148
racesite | -1.250942 .5386628 -2.32 0.020 -2.306702 -.1951827
_cons | -7.79976 1.299528 -6.00 0.000 -10.34679 -5.252732
------------------------------------------------------------------------------
* Stata 8 code.
lfit
* Stata 9 code and output.
estat gof
Logistic model for dfree, goodness-of-fit test
number of observations = 570
number of covariate patterns = 517
Pearson chi2(506) = 482.63
Prob > chi2 = 0.7658
ldev
(5 missing values generated)
(5 missing values generated)
(5 missing values generated)
Logistic model deviance goodness-of-fit test
number of observations = 570
number of covariate patterns = 517
deviance goodness-of-fit = 511.11
degrees of freedom = 506
Prob > chi2 = 0.4282
* Stata 8 code.
lfit, group(10) table
* Stata 9 code and output.
estat gof, group(10) table
Logistic model for dfree, goodness-of-fit test
(Table collapsed on quantiles of estimated probabilities)
_Group _Prob _Obs_1 _Exp_1 _Obs_0 _Exp_0 _Total
1 0.0803 3 3.3 54 53.7 57
2 0.1156 6 5.5 51 51.5 57
3 0.1521 6 7.7 51 49.3 57
4 0.1897 10 9.7 47 47.3 57
5 0.2293 15 12.0 42 45.0 57
6 0.2712 11 14.1 46 42.9 57
7 0.3177 23 16.7 34 40.3 57
8 0.3749 20 19.7 37 37.3 57
9 0.4547 22 23.5 35 33.5 57
10 0.7637 26 29.6 31 27.4 57
number of observations = 570
number of groups = 10
Hosmer-Lemeshow chi2(8) = 6.86
Prob > chi2 = 0.5523
Table 5.10, page 189.
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
Iteration 0: log likelihood = -326.86446
Iteration 1: log likelihood = -300.06724
Iteration 2: log likelihood = -298.98837
Iteration 3: log likelihood = -298.98146
Iteration 4: log likelihood = -298.98146
Logit estimates Number of obs = 575
LR chi2(10) = 55.77
Prob > chi2 = 0.0000
Log likelihood = -298.98146 Pseudo R2 = 0.0853
------------------------------------------------------------------------------
dfree | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .1166385 .0288749 4.04 0.000 .0600446 .1732323
ndrgfp1 | 1.669035 .407152 4.10 0.000 .871032 2.467038
ndrgfp2 | .4336886 .1169052 3.71 0.000 .2045586 .6628185
ivhx2 | -.6346307 .2987192 -2.12 0.034 -1.220109 -.0491518
ivhx3 | -.7049475 .2615805 -2.69 0.007 -1.217636 -.1922591
race | .6841068 .2641355 2.59 0.010 .1664107 1.201803
treat | .4349255 .2037596 2.13 0.033 .035564 .834287
site | .516201 .2548881 2.03 0.043 .0166295 1.015773
agendrgfp1 | -.0152697 .0060268 -2.53 0.011 -.0270819 -.0034575
racesite | -1.429457 .5297806 -2.70 0.007 -2.467808 -.3911062
_cons | -6.843864 1.219316 -5.61 0.000 -9.23368 -4.454048
------------------------------------------------------------------------------
Table 5.11, page 190.
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite, or
Iteration 0: log likelihood = -326.86446
Iteration 1: log likelihood = -300.06724
Iteration 2: log likelihood = -298.98837
Iteration 3: log likelihood = -298.98146
Iteration 4: log likelihood = -298.98146
Logit estimates Number of obs = 575
LR chi2(10) = 55.77
Prob > chi2 = 0.0000
Log likelihood = -298.98146 Pseudo R2 = 0.0853
------------------------------------------------------------------------------
dfree | Odds Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | 1.123713 .0324472 4.04 0.000 1.061884 1.189142
ndrgfp1 | 5.307045 2.160774 4.10 0.000 2.389375 11.78749
ndrgfp2 | 1.542938 .1803775 3.71 0.000 1.226983 1.940253
ivhx2 | .5301313 .1583604 -2.12 0.034 .2951979 .9520366
ivhx3 | .4941345 .129256 -2.69 0.007 .2959289 .825093
race | 1.982001 .5235167 2.59 0.010 1.181058 3.326108
treat | 1.544848 .3147776 2.13 0.033 1.036204 2.303171
site | 1.67565 .4271033 2.03 0.043 1.016768 2.761496
agendrgfp1 | .9848463 .0059354 -2.53 0.011 .9732815 .9965485
racesite | .2394389 .1268501 -2.70 0.007 .0847705 .6763083
------------------------------------------------------------------------------
Table 5.12, page 192.
quietly logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite
lincom 1*race, or
( 1) race = 0.0
------------------------------------------------------------------------------
dfree | Odds Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | 1.982001 .5235167 2.59 0.010 1.181058 3.326108
------------------------------------------------------------------------------
lincom 1*race+1*racesite, or
( 1) race + racesite = 0.0
------------------------------------------------------------------------------
dfree | Odds Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | .474568 .2200132 -1.61 0.108 .1912825 1.177394
------------------------------------------------------------------------------
Figure 5.9, page 194.
NOTE: We were able to get estimates but not the confidence intervals.
logit dfree age ndrgfp1 ndrgfp2 ivhx2 ivhx3 race treat site agendrgfp1 racesite, or
Iteration 0: log likelihood = -326.86446
Iteration 1: log likelihood = -300.06724
Iteration 2: log likelihood = -298.98837
Iteration 3: log likelihood = -298.98146
Iteration 4: log likelihood = -298.98146
Logit estimates Number of obs = 575
LR chi2(10) = 55.77
Prob > chi2 = 0.0000
Log likelihood = -298.98146 Pseudo R2 = 0.0853
------------------------------------------------------------------------------
dfree | Odds Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | 1.123713 .0324472 4.04 0.000 1.061884 1.189142
ndrgfp1 | 5.307045 2.160774 4.10 0.000 2.389375 11.78749
ndrgfp2 | 1.542938 .1803775 3.71 0.000 1.226983 1.940253
ivhx2 | .5301313 .1583604 -2.12 0.034 .2951979 .9520366
ivhx3 | .4941345 .129256 -2.69 0.007 .2959289 .825093
race | 1.982001 .5235167 2.59 0.010 1.181058 3.326108
treat | 1.544848 .3147776 2.13 0.033 1.036204 2.303171
site | 1.67565 .4271033 2.03 0.043 1.016768 2.761496
agendrgfp1 | .9848463 .0059354 -2.53 0.011 .9732815 .9965485
racesite | .2394389 .1268501 -2.70 0.007 .0847705 .6763083
------------------------------------------------------------------------------
NOTE: Below is the code that would be used if we could get the correct confidence intervals.
clear input or se ntreat end serrbar or se treat
Figure 5.10, page 197.
NOTE: We have been unable to recreate these graphs.
Figure 5.11, page 199.
NOTE: We have been unable to recreate these graphs.