Table 4.2 on page 97 using the whas100 dataset.
use https://stats.idre.ucla.edu/stat/examples/asa2/whas100, clear
stset foltime, fail(folstatus)
stcox gender, nohr
failure _d: folstatus
analysis time _t: foltime
Iteration 0: log likelihood = -209.11972
Iteration 1: log likelihood = -207.2544
Iteration 2: log likelihood = -207.2423
Iteration 3: log likelihood = -207.2423
Refining estimates:
Iteration 0: log likelihood = -207.2423
Cox regression -- Breslow method for ties
No. of subjects = 100 Number of obs = 100
No. of failures = 51
Time at risk = 150540
LR chi2(1) = 3.75
Log likelihood = -207.2423 Prob > chi2 = 0.0527
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
gender | .5554704 .2823858 1.97 0.049 .0020044 1.108936
------------------------------------------------------------------------------
Table 4.3 on page 98 using the actg320 dataset.
use https://stats.idre.ucla.edu/stat/examples/asa2/actg320, clear
stset time, fail(censor)
stcox tx, nohr
failure _d: censor
analysis time _t: time
Iteration 0: log likelihood = -658.46549
Iteration 1: log likelihood = -653.12286
Iteration 2: log likelihood = -653.11789
Iteration 3: log likelihood = -653.11789
Refining estimates:
Iteration 0: log likelihood = -653.11789
Cox regression -- Breslow method for ties
No. of subjects = 1151 Number of obs = 1151
No. of failures = 96
Time at risk = 264941
LR chi2(1) = 10.70
Log likelihood = -653.11789 Prob > chi2 = 0.0011
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
tx | -.6843186 .2149187 -3.18 0.001 -1.105551 -.2630857
------------------------------------------------------------------------------
Table 4.4 on page 99 using the whas100 dataset. We use the groups command, which is user written. To install it, type "ssc install groups" in your command window.
use https://stats.idre.ucla.edu/stat/examples/asa2/whas100, clear
stset foltime, fail(folstatus)
recode age 32/59=1 60/69=2 70/79=3 80/92=4, gen(agecat)
tabulate agecat, gen(age)
RECODE of |
age | Freq. Percent Cum.
------------+-----------------------------------
1 | 25 25.00 25.00
2 | 23 23.00 48.00
3 | 22 22.00 70.00
4 | 30 30.00 100.00
------------+-----------------------------------
Total | 100 100.00
groups agecat age2 age3 age4 /* search groups */
+-----------------------------------------------+
| agecat age2 age3 age4 Freq. Percent |
|-----------------------------------------------|
| 1 0 0 0 25 25.00 |
| 2 1 0 0 23 23.00 |
| 3 0 1 0 22 22.00 |
| 4 0 0 1 30 30.00 |
+-----------------------------------------------+
Table 4.5 on page 101 continuing to use the whas100 dataset.
stcox age2 age3 age4, nohr
failure _d: folstatus
analysis time _t: foltime
Iteration 0: log likelihood = -209.11972
Iteration 1: log likelihood = -201.55526
Iteration 2: log likelihood = -201.4586
Iteration 3: log likelihood = -201.45851
Refining estimates:
Iteration 0: log likelihood = -201.45851
Cox regression -- Breslow method for ties
No. of subjects = 100 Number of obs = 100
No. of failures = 51
Time at risk = 150540
LR chi2(3) = 15.32
Log likelihood = -201.45851 Prob > chi2 = 0.0016
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age2 | .0468675 .5186444 0.09 0.928 -.9696568 1.063392
age3 | .9856007 .4453697 2.21 0.027 .112692 1.858509
age4 | 1.262993 .4155407 3.04 0.002 .4485481 2.077438
------------------------------------------------------------------------------
Table 4.6 on page 102 continuing to use the whas100 dataset.
stcox age2 age3 age4
failure _d: folstatus
analysis time _t: foltime
Iteration 0: log likelihood = -209.11972
Iteration 1: log likelihood = -201.55526
Iteration 2: log likelihood = -201.4586
Iteration 3: log likelihood = -201.45851
Refining estimates:
Iteration 0: log likelihood = -201.45851
Cox regression -- Breslow method for ties
No. of subjects = 100 Number of obs = 100
No. of failures = 51
Time at risk = 150540
LR chi2(3) = 15.32
Log likelihood = -201.45851 Prob > chi2 = 0.0016
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age2 | 1.047983 .5435306 0.09 0.928 .3792132 2.896178
age3 | 2.679421 1.193333 2.21 0.027 1.119287 6.414168
age4 | 3.535989 1.469347 3.04 0.002 1.566037 7.983985
------------------------------------------------------------------------------
Table 4.7 on page 102 continuing to use the whas100 dataset.
quietly stcox age2 age3 age4, nohr
matrix list e(V), nohalf format(%6.4f)
symmetric e(V)[3,3]
age2 age3 age4
age2 0.2690 0.1260 0.1251
age3 0.1260 0.1984 0.1260
age4 0.1251 0.1260 0.1727
Table 4.8 on page 105 continuing to use the whas100 dataset. Here, we code the variables using deviations from their means.
foreach var in age2 age3 age4 {
replace `var'=-1 if agecat==1
}
stcox age2 age3 age4, nohr
failure _d: folstatus
analysis time _t: foltime
Iteration 0: log likelihood = -209.11972
Iteration 1: log likelihood = -201.55526
Iteration 2: log likelihood = -201.4586
Iteration 3: log likelihood = -201.45851
Refining estimates:
Iteration 0: log likelihood = -201.45851
Cox regression -- Breslow method for ties
No. of subjects = 100 Number of obs = 100
No. of failures = 51
Time at risk = 150540
LR chi2(3) = 15.32
Log likelihood = -201.45851 Prob > chi2 = 0.0016
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age2 | -.5269978 .3099716 -1.70 0.089 -1.134531 .0805355
age3 | .4117354 .2455778 1.68 0.094 -.0695883 .8930591
age4 | .6891276 .2188689 3.15 0.002 .2601526 1.118103
------------------------------------------------------------------------------
Table 4.9 on page 107 continuing to use the whas100 dataset.
stcox age, nohr
failure _d: folstatus
analysis time _t: foltime
Iteration 0: log likelihood = -209.11972
Iteration 1: log likelihood = -200.64424
Iteration 2: log likelihood = -200.44425
Iteration 3: log likelihood = -200.44391
Refining estimates:
Iteration 0: log likelihood = -200.44391
Cox regression -- Breslow method for ties
No. of subjects = 100 Number of obs = 100
No. of failures = 51
Time at risk = 150540
LR chi2(1) = 17.35
Log likelihood = -200.44391 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .0456618 .0119506 3.82 0.000 .0222391 .0690844
------------------------------------------------------------------------------
Table 4.10 on page 107 using the actg320 dataset.
use https://stats.idre.ucla.edu/stat/examples/asa2/actg320, clear
stset time, fail(censor)
stcox cd4, nohr
failure _d: censor
analysis time _t: time
Iteration 0: log likelihood = -658.46549
Iteration 1: log likelihood = -629.94428
Iteration 2: log likelihood = -626.73801
Iteration 3: log likelihood = -626.63616
Iteration 4: log likelihood = -626.636
Refining estimates:
Iteration 0: log likelihood = -626.636
Cox regression -- Breslow method for ties
No. of subjects = 1151 Number of obs = 1151
No. of failures = 96
Time at risk = 264941
LR chi2(1) = 63.66
Log likelihood = -626.636 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
cd4 | -.0161921 .0025026 -6.47 0.000 -.0210971 -.0112872
------------------------------------------------------------------------------
Table 4.11 on page 113 using the gbcs dataset.
use https://stats.idre.ucla.edu/stat/examples/asa2/gbcs, clear
stset rectime, fail(censrec)
generate hormonexsize = hormone*size
stcox hormone, nohr
failure _d: censrec
analysis time _t: rectime
Iteration 0: log likelihood = -1788.1731
Iteration 1: log likelihood = -1783.774
Iteration 2: log likelihood = -1783.765
Iteration 3: log likelihood = -1783.765
Refining estimates:
Iteration 0: log likelihood = -1783.765
Cox regression -- Breslow method for ties
No. of subjects = 686 Number of obs = 686
No. of failures = 299
Time at risk = 771400
LR chi2(1) = 8.82
Log likelihood = -1783.765 Prob > chi2 = 0.0030
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
hormone | -.3638988 .1250441 -2.91 0.004 -.6089808 -.1188167
------------------------------------------------------------------------------
stcox hormone size, nohr
failure _d: censrec
analysis time _t: rectime
Iteration 0: log likelihood = -1788.1731
Iteration 1: log likelihood = -1776.0514
Iteration 2: log likelihood = -1775.6959
Iteration 3: log likelihood = -1775.6954
Refining estimates:
Iteration 0: log likelihood = -1775.6954
Cox regression -- Breslow method for ties
No. of subjects = 686 Number of obs = 686
No. of failures = 299
Time at risk = 771400
LR chi2(2) = 24.96
Log likelihood = -1775.6954 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
hormone | -.3734745 .125176 -2.98 0.003 -.6188149 -.1281341
size | .0152507 .0035639 4.28 0.000 .0082657 .0222358
------------------------------------------------------------------------------
stcox hormone size hormonexsize, nohr
failure _d: censrec
analysis time _t: rectime
Iteration 0: log likelihood = -1788.1731
Iteration 1: log likelihood = -1776.1541
Iteration 2: log likelihood = -1775.6918
Iteration 3: log likelihood = -1775.6912
Iteration 4: log likelihood = -1775.6912
Refining estimates:
Iteration 0: log likelihood = -1775.6912
Cox regression -- Breslow method for ties
No. of subjects = 686 Number of obs = 686
No. of failures = 299
Time at risk = 771400
LR chi2(3) = 24.96
Log likelihood = -1775.6912 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
hormone | -.3941755 .2601893 -1.51 0.130 -.9041372 .1157862
size | .0149727 .0047047 3.18 0.001 .0057515 .0241938
hormonexsize | .0006522 .0071812 0.09 0.928 -.0134227 .0147272
------------------------------------------------------------------------------
Table 4.12 on page 114 using the uis dataset.
use https://stats.idre.ucla.edu/stat/examples/asa2/uis, clear
stset time, fail(censor)
drop if age==.
drop if ivhx==.
generate drug = ivhx==2 | ivhx==3
generate drugxage = drug*age
stcox drug, nohr
failure _d: censor
analysis time _t: time
Iteration 0: log likelihood = -2831.8568
Iteration 1: log likelihood = -2825.9716
Iteration 2: log likelihood = -2825.9664
Refining estimates:
Iteration 0: log likelihood = -2825.9664
Cox regression -- Breslow method for ties
No. of subjects = 605 Number of obs = 605
No. of failures = 489
Time at risk = 144822
LR chi2(1) = 11.78
Log likelihood = -2825.9664 Prob > chi2 = 0.0006
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
drug | .3210045 .0948107 3.39 0.001 .1351789 .5068301
------------------------------------------------------------------------------
stcox drug age, nohr
failure _d: censor
analysis time _t: time
Iteration 0: log likelihood = -2831.8568
Iteration 1: log likelihood = -2820.2006
Iteration 2: log likelihood = -2820.1989
Refining estimates:
Iteration 0: log likelihood = -2820.1989
Cox regression -- Breslow method for ties
No. of subjects = 605 Number of obs = 605
No. of failures = 489
Time at risk = 144822
LR chi2(2) = 23.32
Log likelihood = -2820.1989 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
drug | .4394466 .1007151 4.36 0.000 .2420487 .6368445
age | -.0263792 .0078392 -3.37 0.001 -.0417438 -.0110147
------------------------------------------------------------------------------
stcox drug age drugxage, nohr
failure _d: censor
analysis time _t: time
Iteration 0: log likelihood = -2831.8568
Iteration 1: log likelihood = -2820.0329
Iteration 2: log likelihood = -2819.8471
Iteration 3: log likelihood = -2819.8471
Iteration 4: log likelihood = -2819.8471
Refining estimates:
Iteration 0: log likelihood = -2819.8471
Cox regression -- Breslow method for ties
No. of subjects = 605 Number of obs = 605
No. of failures = 489
Time at risk = 144822
LR chi2(3) = 24.02
Log likelihood = -2819.8471 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
drug | -.0124116 .5484471 -0.02 0.982 -1.087348 1.062525
age | -.0372282 .0152309 -2.44 0.015 -.0670801 -.0073762
drugxage | .0148441 .0177607 0.84 0.403 -.0199662 .0496543
------------------------------------------------------------------------------
Table 4.13 on page 116 using the whas500 dataset.
use https://stats.idre.ucla.edu/stat/examples/asa2/whas500, clear
stset lenfol, fail(fstat)
generate genderxage = gender*age
stcox gender, nohr
failure _d: fstat
analysis time _t: lenfol
Iteration 0: log likelihood = -1227.579
Iteration 1: log likelihood = -1223.7894
Iteration 2: log likelihood = -1223.7851
Refining estimates:
Iteration 0: log likelihood = -1223.7851
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 441218
LR chi2(1) = 7.59
Log likelihood = -1223.7851 Prob > chi2 = 0.0059
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
gender | .3812532 .137584 2.77 0.006 .1115935 .6509128
------------------------------------------------------------------------------
stcox gender age, nohr
failure _d: fstat
analysis time _t: lenfol
Iteration 0: log likelihood = -1227.579
Iteration 1: log likelihood = -1158.4911
Iteration 2: log likelihood = -1156.574
Iteration 3: log likelihood = -1156.5702
Refining estimates:
Iteration 0: log likelihood = -1156.5702
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 441218
LR chi2(2) = 142.02
Log likelihood = -1156.5702 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
gender | -.0655551 .1405742 -0.47 0.641 -.3410755 .2099653
age | .0668283 .0061941 10.79 0.000 .054688 .0789686
------------------------------------------------------------------------------
stcox gender age genderxage, nohr basehc(h)
failure _d: fstat
analysis time _t: lenfol
Iteration 0: log likelihood = -1227.579
Iteration 1: log likelihood = -1157.5688
Iteration 2: log likelihood = -1153.7302
Iteration 3: log likelihood = -1153.6663
Iteration 4: log likelihood = -1153.6663
Refining estimates:
Iteration 0: log likelihood = -1153.6663
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 441218
LR chi2(3) = 147.83
Log likelihood = -1153.6663 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
gender | 2.328526 .9923413 2.35 0.019 .3835728 4.273479
age | .0784022 .0080213 9.77 0.000 .0626807 .0941236
genderxage | -.0304264 .0125362 -2.43 0.015 -.0549968 -.0058559
------------------------------------------------------------------------------
Figure 4.2 on page 117 continuing to use the whas500 dataset and the model above. The graphs shown on this page use the lean1 scheme.
predict hr1, hr generate lnhr = ln(hr1) twoway (line lnhr age if gender==0)(line lnhr age if gender==1), /// xtitle(Age in Years) ytitle(Estimated Log Hazard) title(Figure 4.2) /// ylabel(2(1)8) legend(order(1 "Males" 2 "Females") ring(0) position(11))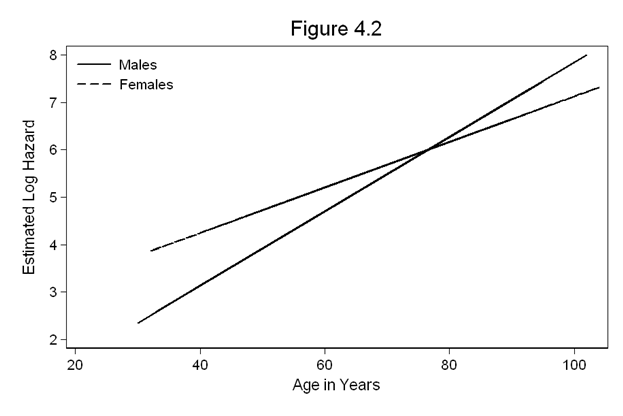
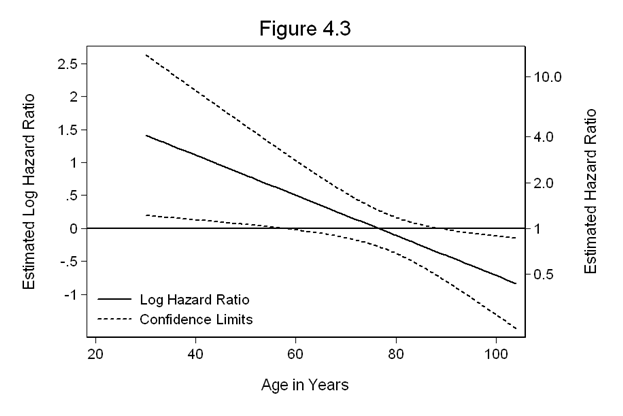
Figure 4.3 on page 118 continuing to use the whas500 dataset. The calculations performed are from equations 4.19-4.22.
matrix V=e(V) gen ln_hr=_b[gender]+_b[genderxage]*age replace hr=exp(ln_hr) gen SE_ln_hr=sqrt(V[1,1]+2*age*V[3,1]+age^2*V[3,3]) gen ln_hr_l=ln_hr-1.96*SE_ln_hr gen ln_hr_u=ln_hr+1.96*SE_ln_hr twoway line ln_hr ln_hr_l ln_hr_u age, sort clpattern(l shortdash shortdash ) /// clcolor(black black black ) /// legend(row(2) col(1) pos(7) order(1 "Log Hazard Ratio" 2 "Confidence Limits") /// ring(0) size(medsmall) region(lc(white)) ) graphregion(color(white)) /// yline(0, lcolor(black)) yaxis(1 2) ytitle(Estimated Log Hazard Ratio) /// ylabel(-1(0.5)2.5, nogrid angle(horizontal)) xscale(titlegap(3)) /// xtitle(Age in Years) ylabel(, axis(2) angle(horizontal)) /// ylabel(-.69 "0.5" 0 "1" .69 "2.0" 1.39 "4.0" 2.30 "10.0", axis(2)) /// ytitle(Estimated Hazard Ratio, axis(2)) /// yscale(titlegap(3) axis(2)) yscale(titlegap(3) axis(1)) /// title(Figure 4.3)
Table 4.14 on page 118 continuing to use the whas500 dataset.
lincom _b[gender]+_b[genderxage]*40, hr
( 1) gender + 40 genderxage = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | 3.038824 1.52132 2.22 0.026 1.139122 8.106639
------------------------------------------------------------------------------
lincom _b[gender]+_b[genderxage]*50, hr
( 1) gender + 50 genderxage = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | 2.241638 .8555832 2.11 0.034 1.060916 4.736419
------------------------------------------------------------------------------
lincom _b[gender]+_b[genderxage]*60, hr
( 1) gender + 60 genderxage = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | 1.653581 .4441904 1.87 0.061 .9767261 2.799484
------------------------------------------------------------------------------
lincom _b[gender]+_b[genderxage]*65, hr
( 1) gender + 65 genderxage = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | 1.420219 .3085033 1.61 0.106 .9278022 2.173979
------------------------------------------------------------------------------
lincom _b[gender]+_b[genderxage]*85, hr
( 1) gender + 85 genderxage = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | .7728143 .124304 -1.60 0.109 .5638492 1.059223
------------------------------------------------------------------------------
lincom _b[gender]+_b[genderxage]*90, hr
( 1) gender + 90 genderxage = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | .6637509 .1330606 -2.04 0.041 .4480915 .9832037
------------------------------------------------------------------------------
Table 4.15 on page 199 using the gbcs dataset.
use https://stats.idre.ucla.edu/stat/examples/asa2/gbcs, clear
stset rectime, fail(censrec)
generate hormonexnodes = hormone*nodes
stcox hormone, nohr
failure _d: censrec
analysis time _t: rectime
Iteration 0: log likelihood = -1788.1731
Iteration 1: log likelihood = -1783.774
Iteration 2: log likelihood = -1783.765
Iteration 3: log likelihood = -1783.765
Refining estimates:
Iteration 0: log likelihood = -1783.765
Cox regression -- Breslow method for ties
No. of subjects = 686 Number of obs = 686
No. of failures = 299
Time at risk = 771400
LR chi2(1) = 8.82
Log likelihood = -1783.765 Prob > chi2 = 0.0030
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
hormone | -.3638988 .1250441 -2.91 0.004 -.6089808 -.1188167
------------------------------------------------------------------------------
stcox hormone nodes, nohr
failure _d: censrec
analysis time _t: rectime
Iteration 0: log likelihood = -1788.1731
Iteration 1: log likelihood = -1759.9643
Iteration 2: log likelihood = -1758.9415
Iteration 3: log likelihood = -1758.9407
Refining estimates:
Iteration 0: log likelihood = -1758.9407
Cox regression -- Breslow method for ties
No. of subjects = 686 Number of obs = 686
No. of failures = 299
Time at risk = 771400
LR chi2(2) = 58.46
Log likelihood = -1758.9407 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
hormone | -.3569371 .1252197 -2.85 0.004 -.6023632 -.111511
nodes | .0576747 .0066566 8.66 0.000 .0446281 .0707213
------------------------------------------------------------------------------
stcox hormone nodes hormonexnodes, nohr
failure _d: censrec
analysis time _t: rectime
Iteration 0: log likelihood = -1788.1731
Iteration 1: log likelihood = -1758.5113
Iteration 2: log likelihood = -1755.9822
Iteration 3: log likelihood = -1755.971
Iteration 4: log likelihood = -1755.971
Refining estimates:
Iteration 0: log likelihood = -1755.971
Cox regression -- Breslow method for ties
No. of subjects = 686 Number of obs = 686
No. of failures = 299
Time at risk = 771400
LR chi2(3) = 64.40
Log likelihood = -1755.971 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
hormone | -.6059966 .1635048 -3.71 0.000 -.9264602 -.285533
nodes | .0492431 .0081727 6.03 0.000 .0332249 .0652614
hormonexno~s | .0381956 .0149761 2.55 0.011 .0088429 .0675483
------------------------------------------------------------------------------
Figure 4.4 on page 120 continuing to use the gbcs dataset. We will use the predicted values and the variance-covariance matrix from the model above to generate the figure.
predict hr1, hr generate lnhr = ln(hr1) matrix V=e(V) gen ln_hr=_b[hormone]+_b[hormonexnodes]*nodes replace hr=exp(ln_hr) gen SE_ln_hr=sqrt(V[1,1]+2*nodes*V[3,1]+nodes^2*V[3,3]) gen ln_hr_l=ln_hr-1.96*SE_ln_hr gen ln_hr_u=ln_hr+1.96*SE_ln_hr twoway line ln_hr ln_hr_l ln_hr_u nodes, sort clpattern(l shortdash shortdash ) /// clcolor(black black black ) /// legend(row(2) col(1) pos(11) order(1 "Log Hazard Ratio" 2 "Confidence Limits") /// ring(0) size(medsmall) region(lc(white)) ) graphregion(color(white)) /// yline(0, lcolor(black)) yaxis(1 2) ytitle(Estimated Log Hazard Ratio) /// ylabel(-1(0.5)2.5, nogrid angle(horizontal)) xscale(titlegap(3)) /// xtitle(Number of Nodes) ylabel(, axis(2) angle(horizontal)) /// ylabel(-.69 "0.5" 0 "1" 1.39 "4.0" 2.71 "15.0", axis(2)) /// ytitle(Estimated Hazard Ratio, axis(2)) /// yscale(titlegap(3) axis(2)) yscale(titlegap(3) axis(1)) /// title(Figure 4.4)
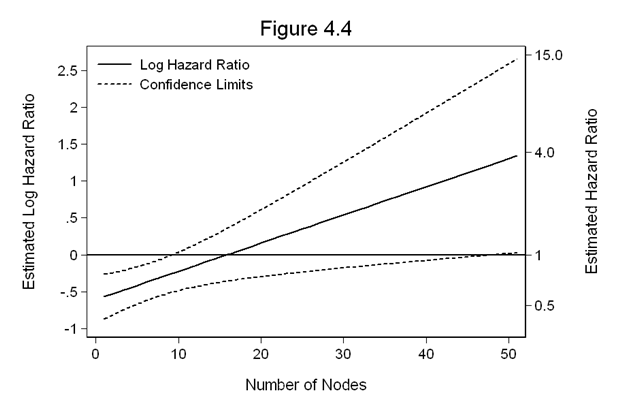
Table 4.16 on page 120 continuing to use the gbcs dataset.
lincom _b[hormone]+_b[hormonexnodes]*1, hr ( 1) hormone + hormonexnodes = 0 ------------------------------------------------------------------------------ _t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- (1) | .5667704 .0874547 -3.68 0.000 .418855 .766921 ------------------------------------------------------------------------------ lincom _b[hormone]+_b[hormonexnodes]*3, hr ( 1) hormone + 3 hormonexnodes = 0 ------------------------------------------------------------------------------ _t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- (1) | .6117633 .085004 -3.54 0.000 .4659183 .8032619 ------------------------------------------------------------------------------ lincom _b[hormone]+_b[hormonexnodes]*5, hr ( 1) hormone + 5 hormonexnodes = 0 ------------------------------------------------------------------------------ _t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- (1) | .660328 .0850732 -3.22 0.001 .512974 .85001 ------------------------------------------------------------------------------ lincom _b[hormone]+_b[hormonexnodes]*7, hr ( 1) hormone + 7 hormonexnodes = 0 ------------------------------------------------------------------------------ _t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- (1) | .712748 .0892621 -2.70 0.007 .557615 .9110402 ------------------------------------------------------------------------------ lincom _b[hormone]+_b[hormonexnodes]*9, hr
( 1) hormone + 9 hormonexnodes = 0 ------------------------------------------------------------------------------ _t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- (1) | .7693293 .0990145 -2.04 0.042 .5978065 .9900654 ------------------------------------------------------------------------------
Table 4.17 on page 121 continuing to use the gbcs dataset.
stcox hormone, nohr failure _d: censrec analysis time _t: rectime Iteration 0: log likelihood = -1788.1731 Iteration 1: log likelihood = -1783.774 Iteration 2: log likelihood = -1783.765 Iteration 3: log likelihood = -1783.765 Refining estimates: Iteration 0: log likelihood = -1783.765 Cox regression -- Breslow method for ties No. of subjects = 686 Number of obs = 686 No. of failures = 299 Time at risk = 771400 LR chi2(1) = 8.82 Log likelihood = -1783.765 Prob > chi2 = 0.0030 ------------------------------------------------------------------------------ _t | Coef. Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- hormone | -.3638988 .1250441 -2.91 0.004 -.6089808 -.1188167 ------------------------------------------------------------------------------
Figure 4.5 on page 123 continuing to use the gbcs dataset.
gen months = rectime / 30.4 stset months, fail(censrec) quietly stcox hormone, nohr basesurv(s45) gen s45_h = s45^(exp(-.364)) sort months twoway line s45 s45_h months, /// legend(row(2) col(1) pos(7) order(1 "No Hormone Therapy" 2 "Hormone Therapy") /// ring(0) size(medsmall)) /// ylabel(0(0.2)1) /// xtitle(Recurrence Time (Months)) /// ytitle(Covariate Adjusted Survival Function) /// title(Figure 4.5)
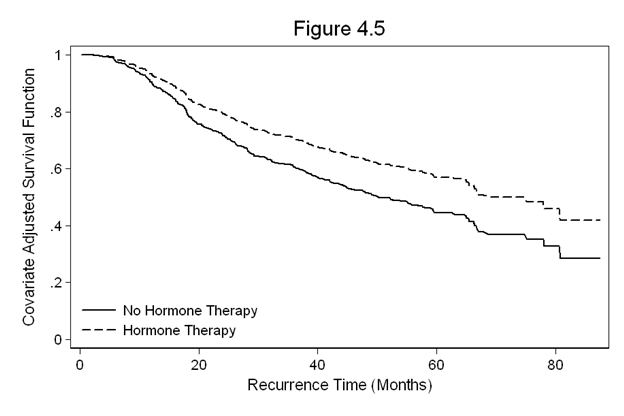
Table 4.18 on page 124 using the gbcs dataset.
stcox hormone size, nohr failure _d: censrec analysis time _t: rectime Iteration 0: log likelihood = -1788.1731 Iteration 1: log likelihood = -1783.774 Iteration 2: log likelihood = -1783.765 Iteration 3: log likelihood = -1783.765 Refining estimates: Iteration 0: log likelihood = -1783.765 Cox regression -- Breslow method for ties No. of subjects = 686 Number of obs = 686 No. of failures = 299 Time at risk = 771400 LR chi2(1) = 8.82 Log likelihood = -1783.765 Prob > chi2 = 0.0030 ------------------------------------------------------------------------------ _t | Coef. Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- hormone | -.3638988 .1250441 -2.91 0.004 -.6089808 -.1188167 ------------------------------------------------------------------------------
Figure 4.6 on page 125 continuing to use the gbcs dataset.
gen size_c = size-25 quietly stcox hormone size_c, nohr basesurv(s46) gen s46_h = s46^(exp(-.373)) sort months twoway line s46 s46_h months, /// legend(row(2) col(1) pos(7) order(1 "No Hormone Therapy" 2 "Hormone Therapy") /// ring(0) size(medsmall)) /// ylabel(0(0.2)1) /// xtitle(Recurrence Time (Months)) /// ytitle(Covariate Adjusted Survival Function) /// title(Figure 4.6)
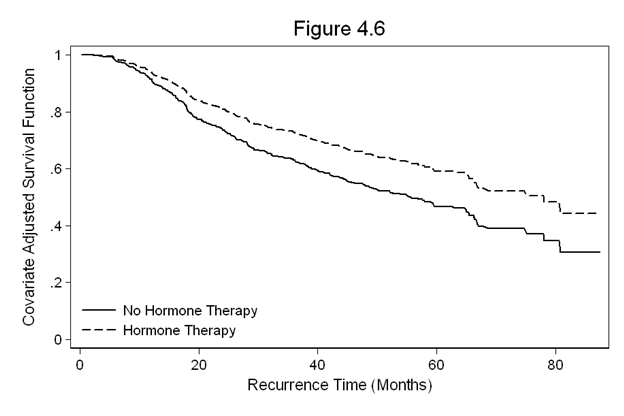
Table 4.19 on page 126 continuing to use the gbcs dataset.
tabulate grade, gen(grade) grade | Freq. Percent Cum. ------------+----------------------------------- 1 | 81 11.81 11.81 2 | 444 64.72 76.53 3 | 161 23.47 100.00 ------------+----------------------------------- Total | 686 100.00 generate ln_prg=ln(prog_recp+1) stcox hormone grade2 grade3 size ln_prg, nohr failure _d: censrec analysis time _t: rectime Iteration 0: log likelihood = -1788.1731 Iteration 1: log likelihood = -1750.0796 Iteration 2: log likelihood = -1748.986 Iteration 3: log likelihood = -1748.9843 Iteration 4: log likelihood = -1748.9843 Refining estimates: Iteration 0: log likelihood = -1748.9843 Cox regression -- Breslow method for ties No. of subjects = 686 Number of obs = 686 No. of failures = 299 Time at risk = 771400 LR chi2(5) = 78.38 Log likelihood = -1748.9843 Prob > chi2 = 0.0000 ------------------------------------------------------------------------------ _t | Coef. Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- hormone | -.3258777 .1261629 -2.58 0.010 -.5731524 -.078603 grade2 | .6244348 .2507907 2.49 0.013 .1328941 1.115975 grade3 | .6289099 .2758348 2.28 0.023 .0882837 1.169536 size | .0135163 .0036485 3.70 0.000 .0063653 .0206673 ln_prg | -.1806934 .0314822 -5.74 0.000 -.2423974 -.1189895 ------------------------------------------------------------------------------
Figure 4.7 on page 127 continuing to use the gbcs dataset.
quietly stcox hormone grade2 grade3 size ln_prg, nohr basesurv(s47) gen s47_10 = s47^(exp(-.487)) gen s47_25 = s47^(exp(-.118)) gen s47_50 = s47^(exp(.239)) gen s47_75 = s47^(exp(.593)) gen s47_90 = s47^(exp(.899)) sort months twoway line s47_10 s47_25 s47_50 s47_75 s47_90 months, /// legend(row(5) col(1) pos(7) order(1 "10th Percentile" 2 "25th Percentile" /// 3 "50th Percentile" 4 "75th Percentile" 5 "90th Percentile") /// ring(0) size(medsmall)) /// ylabel(0(0.2)1) /// xtitle(Recurrence Time (Months)) /// ytitle(Covariate Adjusted Survival Function) /// title(Figure 4.7)
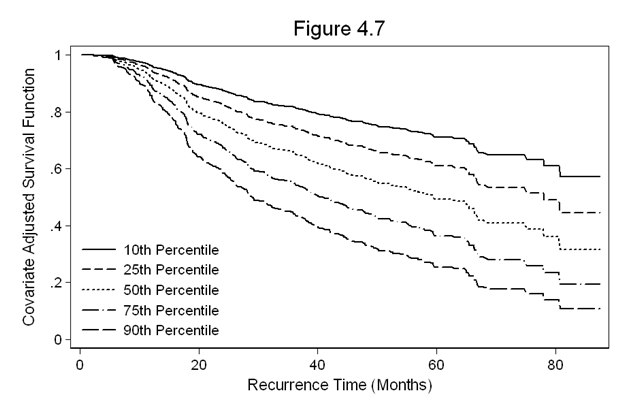
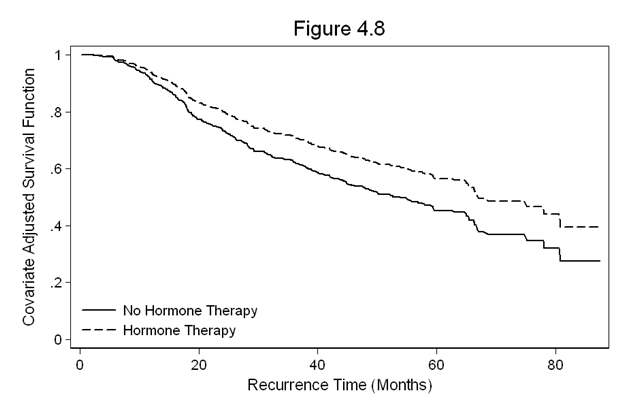
Figure 4.8 on page 129 continuing to use the gbcs dataset.
quietly stcox hormone grade2 grade3 size ln_prg, nohr basesurv(s48) gen s48_noh = s48^(exp(.35)) gen s48_h = s48^(exp(.35-.326)) sort months twoway line s48_noh s48_h months, /// legend(row(2) col(1) pos(7) order(1 "No Hormone Therapy" 2 "Hormone Therapy") /// ring(0) size(medsmall)) /// ylabel(0(0.2)1) /// xtitle(Recurrence Time (Months)) /// ytitle(Covariate Adjusted Survival Function) /// title(Figure 4.8)