Table 5.1 on page 142 using the whas500 data. For each variable, we run separate tests. The numbers seen in the table have been italicized in the output shown here.
use https://stats.idre.ucla.edu/stat/examples/asa2/whas500, clear
generate time = lenfol/365.25
stset time, fail(fstat)
stci, p(50) by(gender)
failure _d: fstat
analysis time _t: time
| no. of
gender | subjects 50% Std. Err. [95% Conf. Interval]
-------------+-------------------------------------------------------------
0 | 300 5.913758 . 4.44627 .
1 | 200 3.605749 .3012859 2.36824 4.32307
-------------+-------------------------------------------------------------
total | 500 4.454483 .4368392 4.1232 6.44216
sts test gender
failure _d: fstat
analysis time _t: time
Log-rank test for equality of survivor functions
| Events Events
gender | observed expected
-------+-------------------------
0 | 111 130.73
1 | 104 84.27
-------+-------------------------
Total | 215 215.00
chi2(1) = 7.79
Pr>chi2 = 0.0053
stci, p(50) by(cvd)
failure _d: fstat
analysis time _t: time
| no. of
cvd | subjects 50% Std. Err. [95% Conf. Interval]
-------------+-------------------------------------------------------------
0 | 125 6.442163 .091912 4.31485 .
1 | 375 4.317591 .4139239 3.72074 6.43395
-------------+-------------------------------------------------------------
total | 500 4.454483 .4368392 4.1232 6.44216
sts test cvd
failure _d: fstat
analysis time _t: time
Log-rank test for equality of survivor functions
| Events Events
cvd | observed expected
------+-------------------------
0 | 45 55.84
1 | 170 159.16
------+-------------------------
Total | 215 215.00
chi2(1) = 2.86
Pr>chi2 = 0.0907
stci, p(50) by(afb)
failure _d: fstat
analysis time _t: time
| no. of
afb | subjects 50% Std. Err. [95% Conf. Interval]
-------------+-------------------------------------------------------------
0 | 422 5.913758 .3102473 4.31485 .
1 | 78 2.368241 .28552 1.14716 3.77002
-------------+-------------------------------------------------------------
total | 500 4.454483 .4368392 4.1232 6.44216
sts test afb
failure _d: fstat
analysis time _t: time
Log-rank test for equality of survivor functions
| Events Events
afb | observed expected
------+-------------------------
0 | 168 184.77
1 | 47 30.23
------+-------------------------
Total | 215 215.00
chi2(1) = 10.90
Pr>chi2 = 0.0010
stci, p(50) by(sho)
failure _d: fstat
analysis time _t: time
| no. of
sho | subjects 50% Std. Err. [95% Conf. Interval]
-------------+-------------------------------------------------------------
0 | 478 5.273101 .4557953 4.20534 .
1 | 22 .0465435 .0029195 .010951 1.22108
-------------+-------------------------------------------------------------
total | 500 4.454483 .4368392 4.1232 6.44216
sts test sho
failure _d: fstat
analysis time _t: time
Log-rank test for equality of survivor functions
| Events Events
sho | observed expected
------+-------------------------
0 | 198 209.33
1 | 17 5.67
------+-------------------------
Total | 215 215.00
chi2(1) = 23.84
Pr>chi2 = 0.0000
stci, p(50) by(chf)
failure _d: fstat
analysis time _t: time
| no. of
chf | subjects 50% Std. Err. [95% Conf. Interval]
-------------+-------------------------------------------------------------
0 | 345 6.455852 .112291 5.91376 .
1 | 155 .9828885 .1189753 .709103 1.53867
-------------+-------------------------------------------------------------
total | 500 4.454483 .4368392 4.1232 6.44216
sts test chf
failure _d: fstat
analysis time _t: time
Log-rank test for equality of survivor functions
| Events Events
chf | observed expected
------+-------------------------
0 | 105 162.38
1 | 110 52.62
------+-------------------------
Total | 215 215.00
chi2(1) = 84.60
Pr>chi2 = 0.0000
stci, p(50) by(av3)
failure _d: fstat
analysis time _t: time
| no. of
av3 | subjects 50% Std. Err. [95% Conf. Interval]
-------------+-------------------------------------------------------------
0 | 489 4.454483 .444643 4.1232 .
1 | 11 5.349761 .5643151 .005476 .
-------------+-------------------------------------------------------------
total | 500 4.454483 .4368392 4.1232 6.44216
sts test av3
failure _d: fstat
analysis time _t: time
Log-rank test for equality of survivor functions
| Events Events
av3 | observed expected
------+-------------------------
0 | 208 210.54
1 | 7 4.46
------+-------------------------
Total | 215 215.00
chi2(1) = 1.52
Pr>chi2 = 0.2171
stci, p(50) by(miord)
failure _d: fstat
analysis time _t: time
| no. of
miord | subjects 50% Std. Err. [95% Conf. Interval]
-------------+-------------------------------------------------------------
0 | 329 5.913758 .1091955 5.2731 6.44216
1 | 171 3.373032 .2971829 1.96578 4.31759
-------------+-------------------------------------------------------------
total | 500 4.454483 .4368392 4.1232 6.44216
sts test miord
failure _d: fstat
analysis time _t: time
Log-rank test for equality of survivor functions
| Events Events
miord | observed expected
------+-------------------------
0 | 125 146.06
1 | 90 68.94
------+-------------------------
Total | 215 215.00
chi2(1) = 9.57
Pr>chi2 = 0.0020
stci , p(50) by(mitype)
failure _d: fstat
analysis time _t: time
| no. of
mitype | subjects 50% Std. Err. [95% Conf. Interval]
-------------+-------------------------------------------------------------
0 | 347 3.501711 .3220688 2.60917 4.44627
1 | 153 6.433949 .1684645 6.43395 .
-------------+-------------------------------------------------------------
total | 500 4.454483 .4368392 4.1232 6.44216
sts test mitype
failure _d: fstat
analysis time _t: time
Log-rank test for equality of survivor functions
| Events Events
mitype | observed expected
-------+-------------------------
0 | 169 141.21
1 | 46 73.79
-------+-------------------------
Total | 215 215.00
chi2(1) = 16.18
Pr>chi2 = 0.0001
Table 5.2 on page 143 using the whas500 data. We are running the Cox regressions using the quietly command, and then displaying the output from the lincom that corresponds to the table in the text.
quietly stcox age, nohr nolog
lincom 5*age, hr
( 1) 5 age = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | 1.392687 .0423237 10.90 0.000 1.312157 1.47816
------------------------------------------------------------------------------
quietly stcox hr, nohr nolog
lincom 10*hr, hr
( 1) 10 hr = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | 1.162248 .0312287 5.60 0.000 1.102625 1.225095
------------------------------------------------------------------------------
quietly stcox sysbp, nohr nolog
lincom 10*sysbp, hr
( 1) 10 sysbp = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | .9559761 .0212814 -2.02 0.043 .9151622 .9986101
------------------------------------------------------------------------------
quietly stcox diasbp, nohr nolog
lincom 10*diasbp, hr
( 1) 10 diasbp = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | .8522054 .0279873 -4.87 0.000 .7990794 .9088635
------------------------------------------------------------------------------
quietly stcox bmi, nohr nolog
lincom 5*bmi, hr
( 1) 5 bmi = 0
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | .6118107 .0451 -6.67 0.000 .5295051 .7069096
------------------------------------------------------------------------------
Table 5.3 on page 143 using the whas500 data in a multivariate model.
stcox age hr sysbp diasbp bmi gender cvd afb chf miord mitype, nolog nohr
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(11) = 208.77
Log likelihood = -1123.1942 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .0486034 .0068246 7.12 0.000 .0352274 .0619795
hr | .0104108 .0030766 3.38 0.001 .0043807 .0164408
sysbp | .0003598 .0029925 0.12 0.904 -.0055054 .006225
diasbp | -.0105894 .0049118 -2.16 0.031 -.0202164 -.0009624
bmi | -.0437268 .0164443 -2.66 0.008 -.075957 -.0114966
gender | -.2705711 .1457033 -1.86 0.063 -.5561443 .0150021
cvd | .0073416 .1781041 0.04 0.967 -.341736 .3564193
afb | .1280092 .1711795 0.75 0.455 -.2074965 .4635149
chf | .7735398 .149917 5.16 0.000 .4797079 1.067372
miord | .0438507 .1484028 0.30 0.768 -.2470134 .3347149
mitype | -.1641189 .1878532 -0.87 0.382 -.5323045 .2040666
------------------------------------------------------------------------------
Table 5.4 on page 144 using the whas500 data in a multivariate model.
stcox age hr diasbp bmi gender afb chf miord mitype, nolog nohr
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(9) = 208.75
Log likelihood = -1123.2029 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .0487438 .0067402 7.23 0.000 .0355332 .0619544
hr | .0103141 .0029871 3.45 0.001 .0044595 .0161687
diasbp | -.0101573 .0035359 -2.87 0.004 -.0170876 -.0032269
bmi | -.0435961 .0163254 -2.67 0.008 -.0755934 -.0115988
gender | -.2682873 .1446723 -1.85 0.064 -.5518399 .0152653
afb | .1254387 .1697283 0.74 0.460 -.2072226 .4581
chf | .7758481 .1486089 5.22 0.000 .48458 1.067116
miord | .044752 .1453871 0.31 0.758 -.2402015 .3297055
mitype | -.1691959 .1837834 -0.92 0.357 -.5294048 .191013
------------------------------------------------------------------------------
Table 5.5 on page 144 using the whas500 data in a multivariate model.
stcox age hr diasbp bmi gender afb chf mitype, nolog nohr
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(8) = 208.66
Log likelihood = -1123.2502 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .0486987 .0067245 7.24 0.000 .0355188 .0618785
hr | .0103887 .0029796 3.49 0.000 .0045487 .0162286
diasbp | -.0102109 .0035346 -2.89 0.004 -.0171386 -.0032832
bmi | -.0439615 .0162951 -2.70 0.007 -.0758994 -.0120236
gender | -.2735672 .1437035 -1.90 0.057 -.5552208 .0080865
afb | .1243358 .1697183 0.73 0.464 -.2083059 .4569776
chf | .7773988 .1486096 5.23 0.000 .4861293 1.068668
mitype | -.1820374 .1789234 -1.02 0.309 -.5327207 .168646
------------------------------------------------------------------------------
Table 5.6 on page 145 using the whas500 data in a multivariate model.
stcox age hr diasbp bmi gender chf, nolog nohr
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(6) = 207.16
Log likelihood = -1123.9997 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .0497904 .006596 7.55 0.000 .0368625 .0627184
hr | .0111788 .0029156 3.83 0.000 .0054644 .0168933
diasbp | -.0105912 .0035124 -3.02 0.003 -.0174753 -.0037071
bmi | -.0451558 .0162751 -2.77 0.006 -.0770543 -.0132573
gender | -.2699895 .1436178 -1.88 0.060 -.5514751 .0114962
chf | .777637 .1466956 5.30 0.000 .4901189 1.065155
------------------------------------------------------------------------------
Table 5.7 on page 146 continuing to use the whas500 data.
summarize age, detail
age
-------------------------------------------------------------
Percentiles Smallest
1% 35.5 30
5% 46 32
10% 49 32 Obs 500
25% 59 33 Sum of Wgt. 500
50% 72 Mean 69.846
Largest Std. Dev. 14.49146
75% 82 97
90% 87 98 Variance 210.0023
95% 90 102 Skewness -.3794081
99% 95 104 Kurtosis 2.372922
/* Calculating Quartile Midpoints for Age */
display (r(min)+r(p25))/2
44.5
display (r(p25)+1+r(p50))/2
66
display (r(p50)+1+r(p75))/2
77.5
display (r(p75)+1+r(max))/2
93.5
gen age_q=age
recode age_q 30/59=1 60/72=2 73/82=3 83/104=4
/* Calculating Coefficients for Age */
xi: stcox i.age_q hr diasbp bmi gender chf, nolog nohr
i.age_q _Iage_q_1-4 (naturally coded; _Iage_q_1 omitted)
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(8) = 205.02
Log likelihood = -1125.0675 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
_Iage_q_2 | .689723 .2944466 2.34 0.019 .1126183 1.266828
_Iage_q_3 | 1.428489 .273315 5.23 0.000 .8928017 1.964177
_Iage_q_4 | 1.807118 .2781617 6.50 0.000 1.261931 2.352305
hr | .0099286 .0029392 3.38 0.001 .0041679 .0156892
diasbp | -.0112784 .0034908 -3.23 0.001 -.0181203 -.0044364
bmi | -.0493604 .0162702 -3.03 0.002 -.0812493 -.0174714
gender | -.2991236 .144762 -2.07 0.039 -.5828519 -.0153953
chf | .833958 .1460775 5.71 0.000 .5476515 1.120265
------------------------------------------------------------------------------
summarize hr, detail
hr
-------------------------------------------------------------
Percentiles Smallest
1% 42 35
5% 54 36
10% 59 36 Obs 500
25% 69 38 Sum of Wgt. 500
50% 85 Mean 87.018
Largest Std. Dev. 23.58623
75% 100.5 154
90% 117 157 Variance 556.3103
95% 128.5 160 Skewness .5650649
99% 150 186 Kurtosis 3.455084
/* Calculating Quartile Midpoints for HR */
display (r(min)+r(p25))/2
52
display (r(p25)+1+r(p50))/2
77.5
display (r(p50)-0.5+r(p75))/2
92.5
display (r(p75)+0.5+r(max))/2
143.5
gen hr_q=hr
recode hr_q 35/69=1 70/85=2 86/100.5=3 100.6/186=4
/* Calculating Coefficients for HR */
xi: stcox age i.hr_q diasbp bmi gender chf, nolog nohr
i.hr_q _Ihr_q_1-4 (naturally coded; _Ihr_q_1 omitted)
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(8) = 209.47
Log likelihood = -1122.8425 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .0504863 .0066777 7.56 0.000 .0373982 .0635744
_Ihr_q_2 | .334802 .2370135 1.41 0.158 -.129736 .79934
_Ihr_q_3 | .4828416 .2122658 2.27 0.023 .0668082 .898875
_Ihr_q_4 | .80903 .2078297 3.89 0.000 .4016912 1.216369
diasbp | -.0100241 .0035105 -2.86 0.004 -.0169046 -.0031436
bmi | -.0458672 .0162975 -2.81 0.005 -.0778097 -.0139247
gender | -.2691365 .1439823 -1.87 0.062 -.5513366 .0130637
chf | .7502223 .1487723 5.04 0.000 .4586339 1.041811
------------------------------------------------------------------------------
summarize diasbp, detail
diasbp
-------------------------------------------------------------
Percentiles Smallest
1% 26 6
5% 48 11
10% 52 20 Obs 500
25% 63 21 Sum o
f Wgt. 500
50% 79 Mean 78.266
Largest Std. Dev. 21.54529
75% 91.5 138
90% 103 140 Variance 464.1996
95% 110 150 Skewness .3069473
99% 134 198 Kurtosis 4.978975
/* Calculating Quartile Midpoints for diasbp */
display (r(min)+r(p25))/2
34.5
display (r(p25)+1+r(p50))/2
71.5
display (r(p50)+1+r(p75))/2
85.75
display (r(p75)+1+r(max))/2
145.25
gen diasbp_q=diasbp
recode diasbp_q 6/63=1 64/79=2 80/91=3 92/198=4
/* Calculating Coefficients for diasbp */
xi: stcox age hr i.diasbp_q bmi gender chf, nolog nohr
i.diasbp_q _Idiasbp_q_1-4 (naturally coded; _Idiasbp_q_1 omitted)
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(8) = 206.50
Log likelihood = -1124.3303 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .0499355 .006599 7.57 0.000 .0370018 .0628692
hr | .011136 .0029239 3.81 0.000 .0054052 .0168668
_Idiasbp_q_2 | -.3136974 .1769299 -1.77 0.076 -.6604736 .0330787
_Idiasbp_q_3 | -.2980701 .2113169 -1.41 0.158 -.7122437 .1161035
_Idiasbp_q_4 | -.6104616 .2125098 -2.87 0.004 -1.026973 -.19395
bmi | -.0458759 .0164737 -2.78 0.005 -.0781637 -.0135882
gender | -.2525157 .1442736 -1.75 0.080 -.5352866 .0302553
chf | .7687909 .1505416 5.11 0.000 .4737349 1.063847
------------------------------------------------------------------------------
summarize bmi, detail
bmi
-------------------------------------------------------------
Percentiles Smallest
1% 15.42587 13.04546
5% 18.60004 14.59031
10% 20.07724 14.83911 Obs 500
25% 23.2068 14.84283 Sum of Wgt. 500
50% 25.94592 Mean 26.61378
Largest Std. Dev. 5.405655
75% 29.39656 42.13576
90% 34.16071 42.38277 Variance 29.22111
95% 36.86193 42.76594 Skewness .528979
99% 40.89478 44.83886 Kurtosis 3.391798
/* Calculating Quartile Midpoints for bmi */
display (r(min)+r(p25))/2
18.12613
display (r(p25)+r(p50))/2
24.576362
display (r(p50)+r(p75))/2
27.671245
display (r(p75)+r(max))/2
37.117712
generate bmi_q=bmi
recode bmi_q 13/23=1 23/26=2 26/29=3 29/45=4
/* Calculating Coefficients for bmi */
xi: stcox age hr diasbp i.bmi_q gender chf, nolog nohr
i.bmi_q _Ibmi_q_1-4 (naturally coded; _Ibmi_q_1 omitted)
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(8) = 209.19
Log likelihood = -1122.9827 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | .0502594 .0066119 7.60 0.000 .0373004 .0632185
hr | .0114025 .0029193 3.91 0.000 .0056809 .0171241
diasbp | -.0111158 .0035745 -3.11 0.002 -.0181217 -.00411
_Ibmi_q_2 | -.45295 .1755822 -2.58 0.010 -.7970849 -.1088152
_Ibmi_q_3 | -.3151134 .1925495 -1.64 0.102 -.6925034 .0622767
_Ibmi_q_4 | -.5921103 .2224768 -2.66 0.008 -1.028157 -.1560638
gender | -.2520627 .1429792 -1.76 0.078 -.5322968 .0281714
chf | .7763868 .1460653 5.32 0.000 .4901041 1.062669
------------------------------------------------------------------------------
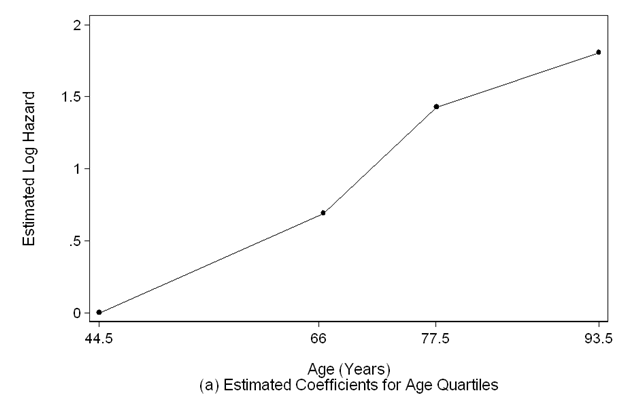
Figure 5.1 on page 146 using the results from Table 5.7.
clear
input agemp age_coeff hrmp hr_coeff diasmp dias_coeff bmimp bmi_coeff
44.55 0.000 52 0.000 34.5 0.000 18.1 0.000
66.5 0.6900 77.5 0.335 71.5 -0.314 24.6 -0.453
77.58 1.428 92.5 0.483 85.8 -0.298 27.7 -0.315
93.5 1.807 143.25 0.809 145.3 -0.610 37.1 -0.592
end
list, clean
agemp age_co~f hrmp hr_coeff diasmp dias_c~f bmimp bmi_co~f
1. 44.55 0 52 0 34.5 0 18.1 0
2. 66.5 .69 77.5 .335 71.5 -.314 24.6 -.453
3. 77.58 1.428 92.5 .483 85.8 -.298 27.7 -.315
4. 93.5 1.807 143.25 .809 145.3 -.61 37.1 -.592
label variable agemp "Age (Years)"
label variable hrmp "Hear Rate (Beats/Min)"
label variable diasmp "Diastolic Blood pressure (mmHg)"
label variable bmimp "Body mass Index (kg/m^2)"
scatter age_coeff agemp if agemp>=44.5 & agemp<=93.5, sort ms(o) c(l) mc(black) ///
clpattern(solid) lc(black) ylabel(,nogrid) clwidth(thin) ///
ytitle(Estimated Log Hazard) yscale(titlegap(3)) xlabel(44.5 66 77.5 93.5) ///
xscale(titlegap(3) ) lc(black) graphregion(fcolor(white)) ///
xtitle("Age (Years)" "(a) Estimated Coefficients for Age Quartiles")
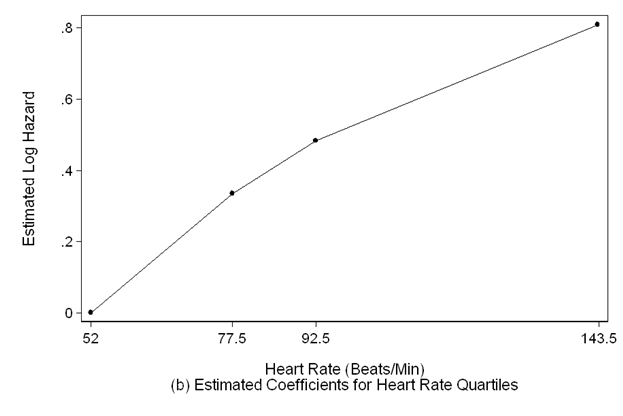
 scatter hr_coeff hrmp if hrmp>=52 &hrmp<=143.5, sort ms(o) c(l) clpattern(solid) ///
lc(black) mc(black) ylabel(,nogrid) clwidth(thin) ///
ytitle(Estimated Log Hazard) yscale(titlegap(3)) xlabel(52 77.5 92.5 143.5) ///
xscale(titlegap(3) ) lc(black) graphregion(fcolor(white)) ///
xtitle("Heart Rate (Beats/Min)" "(b) Estimated Coefficients for Heart Rate Quartiles")
scatter hr_coeff hrmp if hrmp>=52 &hrmp<=143.5, sort ms(o) c(l) clpattern(solid) ///
lc(black) mc(black) ylabel(,nogrid) clwidth(thin) ///
ytitle(Estimated Log Hazard) yscale(titlegap(3)) xlabel(52 77.5 92.5 143.5) ///
xscale(titlegap(3) ) lc(black) graphregion(fcolor(white)) ///
xtitle("Heart Rate (Beats/Min)" "(b) Estimated Coefficients for Heart Rate Quartiles")
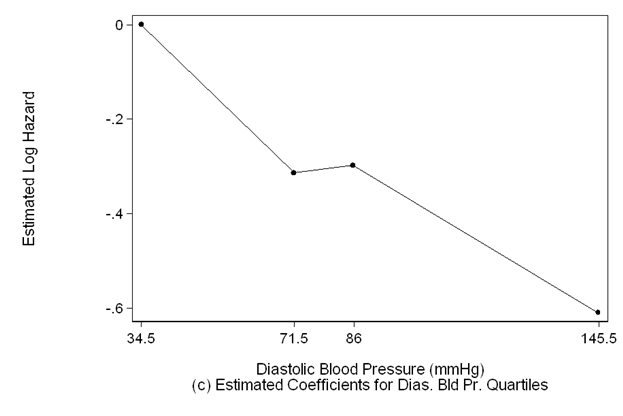
 scatter dias_coeff diasmp if diasmp>=34.5 &diasmp<=145.5, sort ms(o) c(l) clpattern(solid) ///
lc(black) mc(black) ylabel(,nogrid) clwidth(thin) ///
ytitle(Estimated Log Hazard) yscale(titlegap(3)) xlabel(34.5 71.5 86 145.5) ///
xscale(titlegap(3) ) lc(black) graphregion(fcolor(white)) ///
xtitle("Diastolic Blood Pressure (mmHg)" "(c) Estimated Coefficients for Dias. Bld Pr. Quartiles")
scatter dias_coeff diasmp if diasmp>=34.5 &diasmp<=145.5, sort ms(o) c(l) clpattern(solid) ///
lc(black) mc(black) ylabel(,nogrid) clwidth(thin) ///
ytitle(Estimated Log Hazard) yscale(titlegap(3)) xlabel(34.5 71.5 86 145.5) ///
xscale(titlegap(3) ) lc(black) graphregion(fcolor(white)) ///
xtitle("Diastolic Blood Pressure (mmHg)" "(c) Estimated Coefficients for Dias. Bld Pr. Quartiles")
 scatter bmi_coeff bmimp if bmimp>=18.1 &bmimp<=37.1, sort ms(o) c(l) clpattern(solid) lc(black) ///
mc(black) ylabel(,nogrid) clwidth(thin) ///
ytitle(Estimated Log Hazard) yscale(titlegap(3)) xlabel(18.1 24.6 27.7 37.1) ///
xscale(titlegap(3) ) lc(black) graphregion(fcolor(white)) ///
xtitle("Body Mass Index (kg/m^2)" "(d) Estimated Coefficients for Body Mass Index Quartiles")
scatter bmi_coeff bmimp if bmimp>=18.1 &bmimp<=37.1, sort ms(o) c(l) clpattern(solid) lc(black) ///
mc(black) ylabel(,nogrid) clwidth(thin) ///
ytitle(Estimated Log Hazard) yscale(titlegap(3)) xlabel(18.1 24.6 27.7 37.1) ///
xscale(titlegap(3) ) lc(black) graphregion(fcolor(white)) ///
xtitle("Body Mass Index (kg/m^2)" "(d) Estimated Coefficients for Body Mass Index Quartiles")
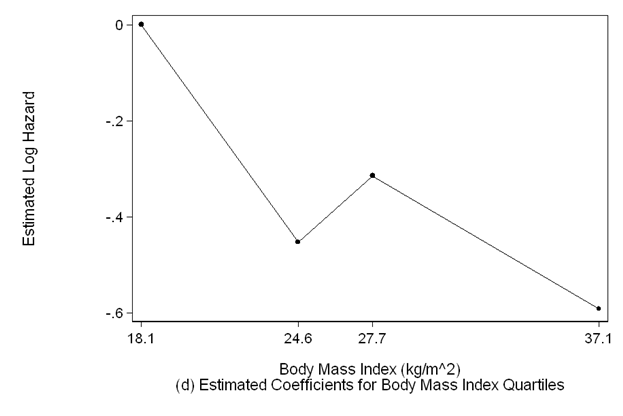
Table 5.8 on page 147 using the whas500 data.
use https://stats.idre.ucla.edu/stat/examples/asa2/whas500, clear
generate time = lenfol/365.25
stset time, fail(fstat)
fracpoly stcox bmi age hr diasbp gender chf, nolog nohr comp log
-> gen double Iage__1 = age-69.846 if e(sample)
-> gen double Ihr__1 = hr-87.018 if e(sample)
-> gen double Idias__1 = diasbp-78.266 if e(sample)
Model # Deviance Power 1 Power 2
1 2241.668 -2.0 .
. . . [additional output omitted for space] . . .
43 2237.784 3.0 2.0
44 2237.922 3.0 3.0
-> gen double Ibmi__1 = X^2-7.082932813 if e(sample)
-> gen double Ibmi__2 = X^3-18.8503615 if e(sample)
(where: X = bmi/10)
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(7) = 217.37
Log likelihood = -1118.8921 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
Ibmi__1 | -.7247551 .172612 -4.20 0.000 -1.063068 -.3864418
Ibmi__2 | .1544204 .0392284 3.94 0.000 .0775343 .2313066
Iage__1 | .0497816 .0066127 7.53 0.000 .0368209 .0627423
Ihr__1 | .0118002 .002954 3.99 0.000 .0060105 .0175899
Idias__1 | -.0106741 .0035015 -3.05 0.002 -.0175368 -.0038113
gender | -.3264025 .1442151 -2.26 0.024 -.609059 -.043746
chf | .82341 .1471777 5.59 0.000 .534947 1.111873
------------------------------------------------------------------------------
Deviance: 2237.78. Best powers of bmi among 44 models fit: 2 3.
Fractional polynomial model comparisons:
---------------------------------------------------------------
bmi df Deviance Dev. dif. P (*) Powers
---------------------------------------------------------------
Not in model 0 2255.945 18.160 0.001
Linear 1 2247.999 10.215 0.017 1
m = 1 2 2241.668 3.884 0.143 -2
m = 2 4 2237.784 -- -- 2 3
---------------------------------------------------------------
(*) P-value from deviance difference comparing reported model with m = 2 model
/* Comparing linear model to model without bmi */
display 2255.945 - 2247.999
7.946
display chi2tail(1,7.946)
.00481938
/* Comparing m = 1 model to linear model */
display 2247.999 - 2241.668
6.331
display chi2tail(1,6.331)
.01186454
/* Comparing m = 2 model to m = 1 model */
display 2241.668 - 2237.784
3.884
display chi2tail(2,3.884)
.14341683
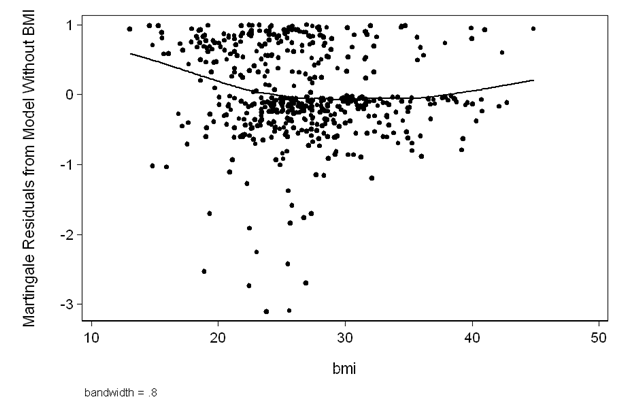
Figure 5.2 on page 149 using the whas500 data.
quietly stcox age hr diasbp gender chf, nolog nohr mgale(M_bmi)
lowess M_bmi bmi, ms(o) ytitle(Martingale Residuals from Model Without BMI) ///
ylabel(,nogrid angle(horizontal)) lineop(lc(black)) mc(black) ///
title("") yscale(titlegap(3)) xscale(titlegap(3)) ///
graphregion(fcolor(white))
 quietly stcox age hr diasbp bmi gender chf, nolog nohr mgale(M_t)
generate Hhat=fstat-M_t
lowess fstat bmi, gen(csm) nograph
lowess Hhat bmi, gen(Hsm) nograph
generate gtfsmooth = ln(csm/Hsm)+_b[bmi]*bmi
twoway line gtfsmooth bmi, sort ytitle("ln(csmoothed/Hsmoothed)+beta*x")
quietly stcox age hr diasbp bmi gender chf, nolog nohr mgale(M_t)
generate Hhat=fstat-M_t
lowess fstat bmi, gen(csm) nograph
lowess Hhat bmi, gen(Hsm) nograph
generate gtfsmooth = ln(csm/Hsm)+_b[bmi]*bmi
twoway line gtfsmooth bmi, sort ytitle("ln(csmoothed/Hsmoothed)+beta*x")
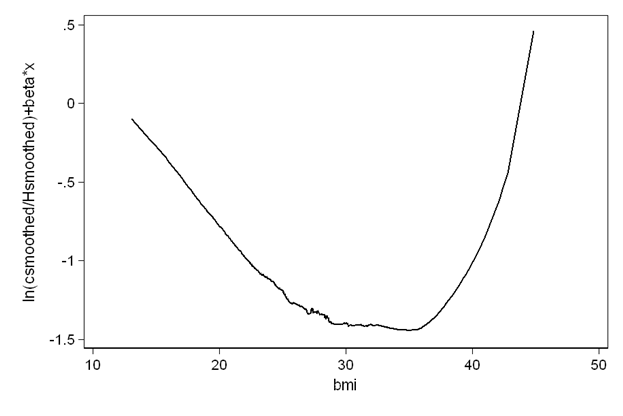
Table 5.9 on page 150 using the whas500 data.
gen bmifp1=(bmi/10)^2
gen bmifp2=(bmi/10)^3
stcox bmifp1 bmifp2 age hr diasbp gender chf, nolog nohr
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(7) = 217.37
Log likelihood = -1118.8921 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
bmifp1 | -.7247552 .172612 -4.20 0.000 -1.063068 -.3864419
bmifp2 | .1544204 .0392284 3.94 0.000 .0775343 .2313066
age | .0497816 .0066127 7.53 0.000 .0368209 .0627423
hr | .0118002 .002954 3.99 0.000 .0060105 .0175899
diasbp | -.0106741 .0035015 -3.05 0.002 -.0175368 -.0038113
gender | -.3264025 .1442151 -2.26 0.024 -.609059 -.043746
chf | .82341 .1471777 5.59 0.000 .534947 1.111873
------------------------------------------------------------------------------
est store maineffects
Figure 5.3 on page 150 using the results from the model above on the whas500 data.
gen fp_funct=_b[bmifp1]*bmifp1+_b[bmifp2]*bmifp2
gen diff=gtfsmooth-fp_funct
quietly summarize diff
replace fp_funct=fp_funct+r(mean)
line gtfsmooth fp_funct bmi, sort clpattern(solid shortdash) ///
legend(row(2) col(1) order(1 "GTF Smooth" 2 "Two Term (2, 3) Model") ring(0) ///
size(medsmall) pos(11) region(lc(white))) ytitle(Estimated Log Hazard) ///
ylabel(,nogrid angle(horizontal)) yscale(titlegap(3)) ///
xscale(titlegap(3)) lc(black black) graphregion(fcolor(white))
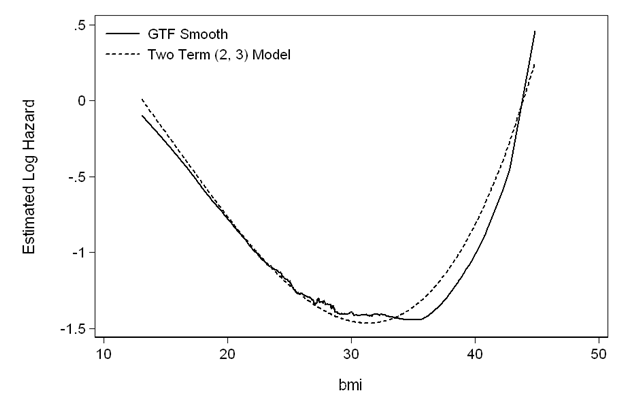
Table 5.10 on page 151 using the whas500 data. Since the BMI test requires removing two variables, we will do these first and then examine the rest of the variables using a foreach loop.
gen bmiage1 = age * bmifp1
gen bmiage2 = age * bmifp2
quietly stcox bmifp1 bmifp2 age hr diasbp gender chf bmiage1 bmiage2, nolog nohr
est store A
quietly stcox bmifp1 bmifp2 age hr diasbp gender chf, nolog nohr
lrtest A
Likelihood-ratio test LR chi2(2) = 3.32
(Assumption: . nested in A) Prob > chi2 = 0.1899
foreach varname of varlist hr diasbp gender chf {
capture drop i1
gen i1=age*(`varname')
quietly stcox bmifp1 bmifp2 age hr diasbp gender chf i1, nolog nohr
est store A
quietly stcox bmifp1 bmifp2 age hr diasbp gender chf, nolog nohr
lrtest A
}
Likelihood-ratio test LR chi2(1) = 2.27
(Assumption: . nested in A) Prob > chi2 = 0.1316
Likelihood-ratio test LR chi2(1) = 2.70
(Assumption: . nested in A) Prob > chi2 = 0.1001
Likelihood-ratio test LR chi2(1) = 5.23
(Assumption: . nested in A) Prob > chi2 = 0.0223
Likelihood-ratio test LR chi2(1) = 4.99
(Assumption: . nested in A) Prob > chi2 = 0.0254
gen bmigender1 = gender * bmifp1
gen bmigender2 = gender * bmifp2
quietly stcox bmifp1 bmifp2 age hr diasbp gender chf bmigender1 bmigender2, nolog nohr
est store A
quietly stcox bmifp1 bmifp2 age hr diasbp gender chf, nolog nohr
lrtest A
Likelihood-ratio test LR chi2(2) = 3.83
(Assumption: . nested in A) Prob > chi2 = 0.1474
foreach varname of varlist hr diasbp chf {
capture drop i1
gen i1=gender*(`varname')
quietly stcox bmifp1 bmifp2 age hr diasbp gender chf i1, nolog nohr
est store A
quietly stcox bmifp1 bmifp2 age hr diasbp gender chf, nolog nohr
lrtest A
}
Likelihood-ratio test LR chi2(1) = 2.13
(Assumption: . nested in A) Prob > chi2 = 0.1446
Likelihood-ratio test LR chi2(1) = 3.07
(Assumption: . nested in A) Prob > chi2 = 0.0796
Likelihood-ratio test LR chi2(1) = 0.50
(Assumption: . nested in A) Prob > chi2 = 0.4817
Table 5.11 on page 152 using the whas500 data.
generate axg = age*gender
stcox bmifp1 bmifp2 age hr diasbp gender chf axg, nolog nohr
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(8) = 222.60
Log likelihood = -1116.2793 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
bmifp1 | -.6730201 .1735831 -3.88 0.000 -1.013237 -.3328035
bmifp2 | .1421257 .0394413 3.60 0.000 .0648222 .2194292
age | .0605347 .0083175 7.28 0.000 .0442327 .0768367
hr | .0116719 .0029427 3.97 0.000 .0059044 .0174394
diasbp | -.0107119 .0034741 -3.08 0.002 -.017521 -.0039027
gender | 1.855257 .9578425 1.94 0.053 -.0220801 3.732594
chf | .823524 .1465418 5.62 0.000 .5363074 1.110741
axg | -.0276658 .0120503 -2.30 0.022 -.051284 -.0040476
------------------------------------------------------------------------------
Table 5.12 on page 152 using the whas500 data. Our coefficient for gender does not match the text exactly.
generate axd = age*diasbp
generate axc = age*chf
generate gxd = gender*diasbp
stcox bmifp1 bmifp2 age hr diasbp gender chf axg axc axd gxd, nolog nohr
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(11) = 229.97
Log likelihood = -1112.5953 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
bmifp1 | -.6410106 .1753848 -3.65 0.000 -.9847585 -.2972627
bmifp2 | .137229 .0400039 3.43 0.001 .0588228 .2156352
age | .0338233 .0203215 1.66 0.096 -.0060061 .0736527
hr | .011193 .0029711 3.77 0.000 .0053697 .0170164
diasbp | -.0526922 .0213007 -2.47 0.013 -.0944408 -.0109435
gender | .7476962 1.220066 0.61 0.540 -1.643588 3.138981
chf | 2.456034 1.00301 2.45 0.014 .4901696 4.421898
axg | -.0220185 .0131616 -1.67 0.094 -.0478148 .0037779
axc | -.020293 .0127172 -1.60 0.111 -.0452182 .0046323
axd | .0004845 .0002649 1.83 0.067 -.0000346 .0010037
gxd | .0089871 .0069291 1.30 0.195 -.0045937 .022568
------------------------------------------------------------------------------
Table 5.13 on page 153 using the whas500 data.
stcox bmifp1 bmifp2 age hr diasbp gender chf axg, nolog nohr
failure _d: fstat
analysis time _t: time
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(8) = 222.60
Log likelihood = -1116.2793 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
bmifp1 | -.6730201 .1735831 -3.88 0.000 -1.013237 -.3328035
bmifp2 | .1421257 .0394413 3.60 0.000 .0648222 .2194292
age | .0605347 .0083175 7.28 0.000 .0442327 .0768367
hr | .0116719 .0029427 3.97 0.000 .0059044 .0174394
diasbp | -.0107119 .0034741 -3.08 0.002 -.017521 -.0039027
gender | 1.855257 .9578425 1.94 0.053 -.0220801 3.732594
chf | .823524 .1465418 5.62 0.000 .5363074 1.110741
axg | -.0276658 .0120503 -2.30 0.022 -.051284 -.0040476
------------------------------------------------------------------------------
Table 5.14 on page 158 using the whas500 data.
Stata computes the Wald test but not the score test and the output is not formatted as in the book. However, we can compare the order in which variables were added to that shown in the book. Stata provides the p-values at each step and the final model.
stepwise stcox age chf hr diasbp bmi gender mitype miord sysbp cvd afb, ///
pe(.25) pr(.8) forward
begin with empty model
p = 0.0000 < 0.2500 adding age
p = 0.0000 < 0.2500 adding chf
p = 0.0025 < 0.2500 adding hr
p = 0.0022 < 0.2500 adding diasbp
p = 0.0074 < 0.2500 adding bmi
p = 0.0601 < 0.2500 adding gender
Cox regression -- Breslow method for ties
No. of subjects = 500 Number of obs = 500
No. of failures = 215
Time at risk = 1207.989046
LR chi2(6) = 207.16
Log likelihood = -1123.9997 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
age | 1.051051 .0069327 7.55 0.000 1.03755 1.064727
chf | 2.176323 .319257 5.30 0.000 1.63251 2.901289
hr | 1.011242 .0029484 3.83 0.000 1.005479 1.017037
diasbp | .9894647 .0034754 -3.02 0.003 .9826765 .9962998
bmi | .9558485 .0155565 -2.77 0.006 .9258395 .9868302
gender | .7633875 .109636 -1.88 0.060 .5760994 1.011563
------------------------------------------------------------------------------
Table 5.15 on page 161 using the whas500 data.
Stata does not have built-in score test or Mallow’s C.
It is possible to compute these values manually using the scoretest_cox command.
We will show the process for the first line in Table 5.15.
To get scoretest_cox type command: search scoretest_cox
/* 11-covariate model */
quietly stcox age gender hr diasbp bmi chf sysbp cvd afb miord mitype
scoretest_cox age gender hr diasbp bmi chf sysbp cvd afb miord mitype
( 1) age = 0
( 2) gender = 0
( 3) hr = 0
( 4) diasbp = 0
( 5) bmi = 0
( 6) chf = 0
( 7) sysbp = 0
( 8) cvd = 0
( 9) afb = 0
(10) miord = 0
(11) mitype = 0
chi2( 11) = 208.62
Prob > chi2 = 0.0000
scalar score0 = r(chi2)
/* Model 1 */
quietly stcox age gender hr diasbp bmi chf
scoretest_cox age gender hr diasbp bmi chf
( 1) age = 0
( 2) gender = 0
( 3) hr = 0
( 4) diasbp = 0
( 5) bmi = 0
( 6) chf = 0
chi2( 6) = 206.49
Prob > chi2 = 0.0000
scalar score1 = r(chi2)
display score0 - score1
2.1326329
display (score0 - score1) + (11-2*5)
3.1326329
/* Model 2 */
quietly stcox age gender hr diasbp bmi chf mitype
scoretest_cox age gender hr diasbp bmi chf mitype
( 1) age = 0
( 2) gender = 0
( 3) hr = 0
( 4) diasbp = 0
( 5) bmi = 0
( 6) chf = 0
( 7) mitype = 0
chi2( 7) = 208.04
Prob > chi2 = 0.0000
scalar score2 = r(chi2)
display score0 - score2
.58451409
display (score0 - score2) + (11-2*4)
3.5845141
/* Model 3 */
quietly stcox age hr diasbp bmi chf
scoretest_cox age hr diasbp bmi chf
( 1) age = 0
( 2) hr = 0
( 3) diasbp = 0
( 4) bmi = 0
( 5) chf = 0
chi2( 5) = 202.67
Prob > chi2 = 0.0000
scalar score3 = r(chi2)
display score0 - score3
5.9477082
display (score0 - score3) + (11-2*6)
4.9477082
/* Model 4 */
quietly stcox age gender hr diasbp bmi chf sysbp
scoretest_cox age gender hr diasbp bmi chf sysbp
( 1) age = 0
( 2) gender = 0
( 3) hr = 0
( 4) diasbp = 0
( 5) bmi = 0
( 6) chf = 0
( 7) sysbp = 0
chi2( 7) = 206.86
Prob > chi2 = 0.0000
scalar score4 = r(chi2)
display score0 - score4
1.7659588
display (score0 - score4) + (11-2*4)
4.7659588
/* Model 5 */
quietly stcox age gender hr diasbp bmi chf miord
scoretest_cox age gender hr diasbp bmi chf miord
( 1) age = 0
( 2) gender = 0
( 3) hr = 0
( 4) diasbp = 0
( 5) bmi = 0
( 6) chf = 0
( 7) miord = 0
chi2( 7) = 206.64
Prob > chi2 = 0.0000
scalar score5 = r(chi2)
display score0 - score5
1.9869457
display (score0 - score5) + (11-2*4)
4.9869457
Table 5.16 on page 165 using the whas500 data. To install the mfp command, type search mfp. We have displayed only the output that corresponds to the table in the text.
mfp stcox age hr sysbp diasbp bmi gender cvd afb chf miord mitype, ///
adjust(no) df(4, gender cvd afb chf miord mitype:1) ///
alpha(0.15) select(0.15)
Deviance for model with all terms untransformed = 2246.388, 500 observations
. . . [additional output omitted for space] . . .
Variable Model (vs.) Deviance Dev diff. P Powers (vs.)
----------------------------------------------------------------------
age null FP2 2301.581 67.477 0.000* . 3 3
lin. 2237.784 3.680 0.298 1
Final 2237.784 1
chf null lin. 2268.808 31.023 0.000* . 1
Final 2237.784 1
hr null FP2 2253.103 18.116 0.001* . -2 -2
lin. 2237.784 2.797 0.424 1
Final 2237.784 1
bmi null FP2 2255.945 18.160 0.001* . 2 3
lin. 2247.999 10.215 0.017+ 1
FP1 2241.668 3.884 0.143+ -2
Final 2237.784 2 3
diasbp null FP2 2247.368 12.275 0.015* . 3 3
lin. 2237.784 2.691 0.442 1
Final 2237.784 1
gender null lin. 2242.906 5.122 0.024* . 1
Final 2237.784 1
mitype null lin. 2237.784 0.852 0.356 . 1
Final 2237.784 .
afb null lin. 2237.784 0.234 0.629 . 1
Final 2237.784 .
miord null lin. 2237.784 0.153 0.695 . 1
Final 2237.784 .
sysbp null FP2 2237.784 3.095 0.542 . -.5 -.5
Final 2237.784 .
cvd null lin. 2237.784 0.052 0.820 . 1
Final 2237.784 .
Fractional polynomial fitting algorithm converged after 3 cycles.
. . . [additional output omitted for space] . . .
Tables 5.17 & 5.18 use hypothetical data
