Table 7.1 on page 211.
use https://stats.idre.ucla.edu/stat/examples/asa2/actg320, clear
stset time, fail(censor)
failure event: censor != 0 & censor < .
obs. time interval: (0, time]
exit on or before: failure
------------------------------------------------------------------------------
1151 total obs.
0 exclusions
------------------------------------------------------------------------------
1151 obs. remaining, representing
96 failures in single record/single failure data
264941 total analysis time at risk, at risk from t = 0
earliest observed entry t = 0
last observed exit t = 364
generate ivdrug_d=ivdrug>1
generate karnof_90=(karnof==90)
generate karnof_70_80=(karnof==70|karnof==80)
centile cd4, c(25 50 75)
-- Binom. Interp. --
Variable | Obs Percentile Centile [95% Conf. Interval]
-------------+-------------------------------------------------------------
cd4 | 1151 25 23 19.5 26.09482
| 50 74.5 67.5 80.5
| 75 136.5 130 141.1559
generate cd4_q=1
replace cd4_q=2 if cd4>23 & cd4<=74.5
replace cd4_q=3 if cd4>74.5 & cd4<=136.5
replace cd4_q=4 if cd4>136.5
stcox tx ivdrug_d karnof_70_80 karnof_90 age, nolog nohr strata(cd4_q) ///
bases(s0)
failure _d: censor
analysis time _t: time
Stratified Cox regr. -- Breslow method for ties
No. of subjects = 1151 Number of obs = 1151
No. of failures = 96
Time at risk = 264941
LR chi2(5) = 36.73
Log likelihood = -506.75521 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
tx | -.6677746 .215533 -3.10 0.002 -1.090212 -.2453376
ivdrug_d | -.5463155 .3225604 -1.69 0.090 -1.178522 .0858912
karnof_70_80 | 1.191 .2962962 4.02 0.000 .6102697 1.77173
karnof_90 | .4118837 .2926676 1.41 0.159 -.1617343 .9855017
age | .0224756 .0112014 2.01 0.045 .0005213 .04443
------------------------------------------------------------------------------
Stratified by cd4_q
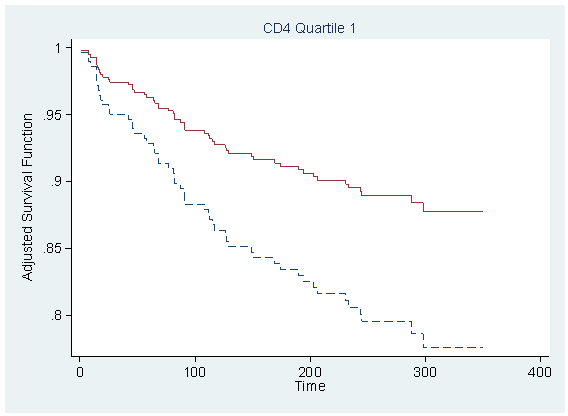
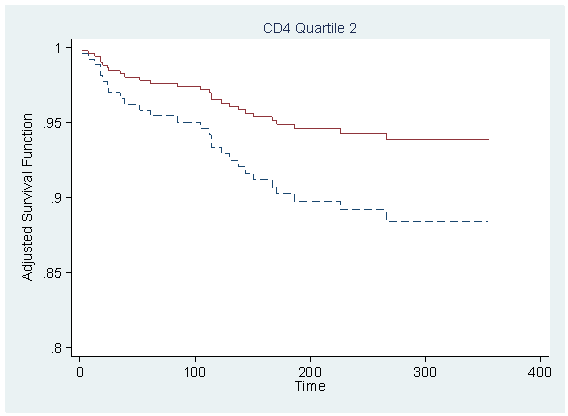
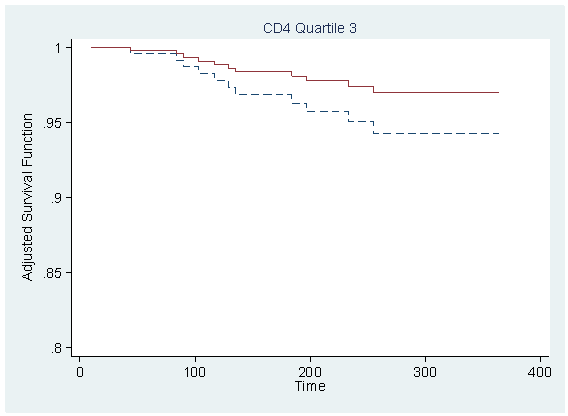
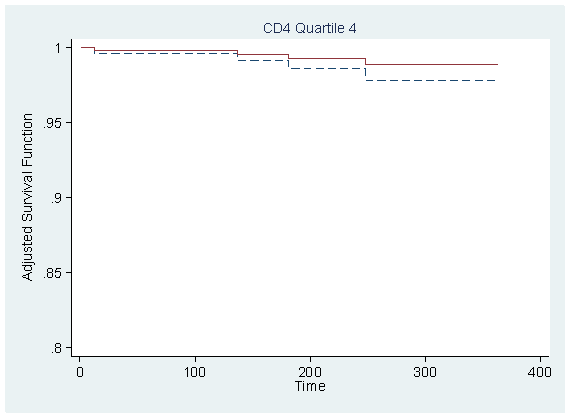
Figure 7.1 on page 212 using the model from previous example.
local tx=_b[tx] predict r,xb replace r = r-tx*_b[tx] table cd4_q, con(median r)
----------------------
cd4_q | med(r)
----------+-----------
1 | 1.217556
2 | 1.176055
3 | 1.086152
4 | 1.093729
----------------------
sort time
foreach i of numlist 1/4 {
quietly sum r if cd4_q==`i', detail
local median = r(p50)
gen s`i' = s0^(exp(`median'))
gen s`i'_tr = s0^(exp(`median'+`tx'))
scatter s`i' s`i'_tr time if cd4_q==`i', sort ms(none none) c(J J) clpattern(dash solid) ///
ylabel(0.8(0.05)1) title( "CD4 Quartile `i'" , size(medsmall) pos(12) ) ///
ytitle(Adjusted Survival Function) ylabel(,nogrid angle(horizontal)) ///
xtitle("Time") legend(off) name(s`i', replace)
}
Table 7.2 on page 217 and Table 7.3 on page 219 using uis data.
use https://stats.idre.ucla.edu/stat/examples/asa2/uis, clear
generate enter=410-los
generate exit=410+(time-los)
stset exit, enter(enter) fail(censor) id(id)
id: id
failure event: censor != 0 & censor < .
obs. time interval: (exit[_n-1], exit]
enter on or after: time enter
exit on or before: failure
------------------------------------------------------------------------------
628 total obs.
0 exclusions
------------------------------------------------------------------------------
628 obs. remaining, representing
628 subjects
508 failures in single failure-per-subject data
147394 total analysis time at risk, at risk from t = 0
earliest observed entry t = 10
last observed exit t = 1531
stsplit off_tx, at(0,410)
replace off_tx=1 if off_tx==410
rename _t oldt
stset oldt, origin(_t0) id(id) fail(censor)
id: id
failure event: censor != 0 & censor < .
obs. time interval: (oldt[_n-1], oldt]
exit on or before: failure
t for analysis: (time-origin)
origin: time oldt0
------------------------------------------------------------------------------
1174 total obs.
0 exclusions
------------------------------------------------------------------------------
1174 obs. remaining, representing
628 subjects
508 failures in single failure-per-subject data
147394 total analysis time at risk, at risk from t = 0
earliest observed entry t = 0
last observed exit t = 1172
xi:stcox i.treat*off_tx, nolog nohr
i.treat _Itreat_0-1 (naturally coded; _Itreat_0 omitted)
i.treat*off_tx _ItreXoff_t_# (coded as above)
failure _d: censor
analysis time _t: (oldt-origin)
origin: time oldt0
id: id
Cox regression -- Breslow method for ties
No. of subjects = 628 Number of obs = 1174
No. of failures = 508
Time at risk = 147394
LR chi2(3) = 387.12
Log likelihood = -2766.9447 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
_Itreat_1 | -.52389 .2258328 -2.32 0.020 -.9665141 -.081266
off_tx | 2.270329 .1865035 12.17 0.000 1.904789 2.635869
_ItreXoff_~1 | .6209768 .2463036 2.52 0.012 .1382306 1.103723
------------------------------------------------------------------------------
Table 7.3
xi:stcox i.treat*off_tx, nolog
i.treat _Itreat_0-1 (naturally coded; _Itreat_0 omitted)
i.treat*off_tx _ItreXoff_t_# (coded as above)
failure _d: censor
analysis time _t: (oldt-origin)
origin: time oldt0
id: id
Cox regression -- Breslow method for ties
No. of subjects = 628 Number of obs = 1174
No. of failures = 508
Time at risk = 147394
LR chi2(3) = 387.12
Log likelihood = -2766.9447 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Haz. Ratio Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
_Itreat_1 | .5922123 .1337409 -2.32 0.020 .3804068 .9219485
off_tx | 9.682585 1.805836 12.17 0.000 6.717989 13.95544
_ItreXoff_~1 | 1.860745 .4583082 2.52 0.012 1.14824 3.015372
------------------------------------------------------------------------------
lincom _Itreat_1 + _ItreXoff_t_1
( 1) _Itreat_1 + _ItreXoff_t_1 = 0
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | .0970868 .097709 0.99 0.320 -.0944193 .2885929
------------------------------------------------------------------------------
di exp( r(estimate) )
1.101956
di exp( r(estimate) + 1.96*r(se))
1.334553
di exp( r(estimate) - 1.96*r(se))
.90989798
lincom off_tx + _ItreXoff_t_1
( 1) off_tx + _ItreXoff_t_1 = 0
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
(1) | 2.891306 .2050128 14.10 0.000 2.489488 3.293123
------------------------------------------------------------------------------
di exp( r(estimate) )
18.01682
di exp( r(estimate) + 1.96*r(se))
26.927036
di exp( r(estimate) - 1.96*r(se))
12.055015
Table 7.5 on page 222 and Table 7.6 on page 223 using grace1000 data.
use https://stats.idre.ucla.edu/stat/examples/asa2/grace1000, clear
stset days, fail(death)
failure event: death != 0 & death < .
obs. time interval: (0, days]
exit on or before: failure
------------------------------------------------------------------------------
1000 total obs.
0 exclusions
------------------------------------------------------------------------------
1000 obs. remaining, representing
324 failures in single record/single failure data
109847.5 total analysis time at risk, at risk from t = 0
earliest observed entry t = 0
last observed exit t = 180
generate age_inv = 1/age*1000
generate sysbp_sqrt = sqrt(sysbp)
stcox revasc age_inv sysbp_sqrt st , nolog nohr
failure _d: death
analysis time _t: days
Cox regression -- Breslow method for ties
No. of subjects = 1000 Number of obs = 1000
No. of failures = 324
Time at risk = 109847.5
LR chi2(4) = 185.82
Log likelihood = -2043.7251 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
revasc | -.5296277 .1171956 -4.52 0.000 -.759327 -.2999285
age_inv | -.1890701 .0224795 -8.41 0.000 -.2331292 -.145011
sysbp_sqrt | -.2072934 .0386913 -5.36 0.000 -.2831269 -.1314599
stchange | .5134163 .1188503 4.32 0.000 .2804741 .7463586
------------------------------------------------------------------------------
Table 7.6
generate enter=200-revascdays
generate exit=200+(days-revascdays)
stset exit, enter(enter) fail(death) id(id)
id: id
failure event: death != 0 & death < .
obs. time interval: (exit[_n-1], exit]
enter on or after: time enter
exit on or before: failure
------------------------------------------------------------------------------
1000 total obs.
0 exclusions
------------------------------------------------------------------------------
1000 obs. remaining, representing
1000 subjects
324 failures in single failure-per-subject data
109847.5 total analysis time at risk, at risk from t = 0
earliest observed entry t = 20
last observed exit t = 380
stsplit revasc_t, at(0,200)
replace revasc_t=1 if revasc_t==200
replace revasc_t=0 if revasc==0
stset _t, origin(_t0) id(id) fail(death)
id: id
failure event: death != 0 & death < .
obs. time interval: (_t[_n-1], _t]
exit on or before: failure
t for analysis: (time-origin)
origin: time _t0
------------------------------------------------------------------------------
1359 total obs.
0 exclusions
------------------------------------------------------------------------------
1359 obs. remaining, representing
1000 subjects
324 failures in single failure-per-subject data
109847.5 total analysis time at risk, at risk from t = 0
earliest observed entry t = 0
last observed exit t = 180
stcox revasc_t age_inv sysbp_sqrt st , nolog nohr
failure _d: death
analysis time _t: (_t-origin)
origin: time _t0
id: id
Cox regression -- Breslow method for ties
No. of subjects = 1000 Number of obs = 1359
No. of failures = 324
Time at risk = 109847.5
LR chi2(4) = 169.38
Log likelihood = -2051.946 Prob > chi2 = 0.0000
------------------------------------------------------------------------------
_t | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
revasc_t | -.2609491 .1237083 -2.11 0.035 -.503413 -.0184853
age_inv | -.2020569 .0228396 -8.85 0.000 -.2468218 -.1572921
sysbp_sqrt | -.2146887 .0391799 -5.48 0.000 -.2914798 -.1378975
stchange | .500509 .1190631 4.20 0.000 .2671496 .7338684
------------------------------------------------------------------------------