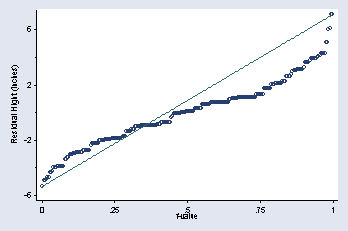
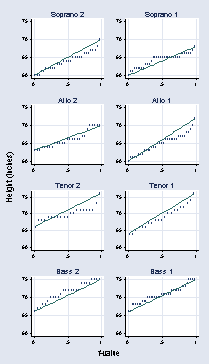
page 16, Figure 2.1
use "c:vizdatasinger.dta", clear
* panel 1
quantile height if voice_part == "Soprano 2", title("Soprano 2") ///
xtitle("") ytitle("") xlabel(0 .5 1, grid) ylabel(60(5)75, angle(0)) ///
xmtick(.25 .75) name(a, replace) msymbol(Oh)
* panel 2
quantile height if voice_part == "Alto 2", title("Alto 2") ///
xtitle("") ytitle("") xlabel(0 .5 1, grid) ylabel(60(5)75, angle(0)) ///
xmtick(.25 .75) name(b, replace) msymbol(Oh)
* panel 3
quantile height if voice_part == "Tenor 2", title("Tenor 2") xtitle("") ///
ytitle("") xlabel(0 .5 1, grid) ylabel(60(5)75, angle(0)) xmtick(.25 .75) ///
name(c, replace) msymbol(Oh)
* panel 4
quantile height if voice_part == "Bass 2", title("Bass 2") xtitle("") ///
ytitle("") xlabel(0 .5 1, grid) ylabel(60(5)75, angle(0)) xmtick(.25 .75) ///
name(d, replace) msymbol(Oh)
* panel 5
quantile height if voice_part == "Soprano 1", title("Soprano 1") ///
xtitle("") ytitle("") xlabel(0 .5 1, grid) ylabel(60(5)75, angle(0)) ///
xmtick(.25 .75) name(e, replace) msymbol(Oh)
* panel 6
quantile height if voice_part == "Alto 1", title("Alto 1") xtitle("") ///
ytitle("") xlabel(0 .5 1, grid) ylabel(60(5)75, angle(0)) xmtick(.25 .75) ///
name(f, replace) msymbol(Oh)
* panel 7
quantile height if voice_part == "Tenor 1", title("Tenor 1") xtitle("") ///
ytitle("") xlabel(0 .5 1, grid) ylabel(60(5)75, angle(0)) xmtick(.25 .75) ///
name(g, replace) msymbol(Oh)
* panel 8
quantile height if voice_part == "Bass 1", title("Bass 1") xtitle("") ///
ytitle("") xlabel(0 .5 1, grid) ylabel(60(5)75, angle(0)) xmtick(.25 .75) ///
name(h, replace) msymbol(Oh)
graph combine a e b f c g d h, b1title("f-value") ///
l1title("Height (inches)") cols(2) xsize(2) ysize(3.6)
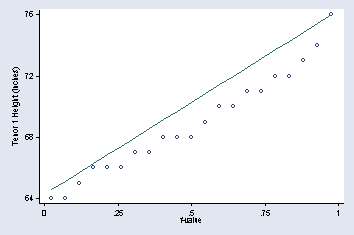
page 19, Figure 2.2
quantile height if voice_part == "Tenor 1", ///
ytitle("Tenor 1 Height (inches)") ylabel(64(4)76, angle(0) nogrid) ///
xtitle("f-value") msymbol(Oh)
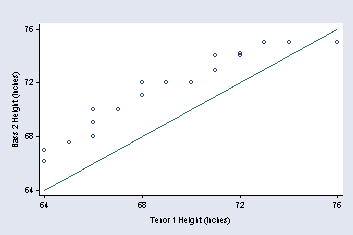
page 22, Figure 2.3
NOTE: The first set of quotes in the xtitle option are used to move the title of the x-axis down a little bit.
gen hbass2 = height if voice_part=="Bass 2"
(209 missing values generated)
gen htenor1 = height if voice_part=="Tenor 1"
(214 missing values generated)
qqplot hbass2 htenor1, ytitle("Bass 2 Height (inches)") msymbol(Oh) ///
ylabel(64(4)76, angle(0) nogrid) xtitle(" " "Tenor 1 Height (inches)") ///
title(" ") xlabel(64(4)76)
page 23, Figure 2.4
NOTE: The tmdplot .ado was written by ATS to create the m-d (mean difference) plot. You can download the command using the following commands:
net from https://stats.idre.ucla.edu/stat/stata/ado/analysis
net install tmdplot
use "c:vizdatasinger.dta", clear
gen hbass2 = height if voice_part=="Bass 2"
(209 missing values generated)
gen htenor1 = height if voice_part=="Tenor 1"
(214 missing values generated)
tmdplot hbass2 htenor1, yline(0) xtitle(Mean (inches)) ///
ytitle(Difference (inches)) ylabel(-1(1)4, nogrid angle(0)) ///
xlabel(66(2)76) msymbol(Oh) title("")
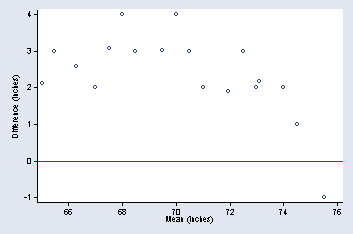
page 24, Figure 2.5
This is a matrix of qqplots and has been skipped for now.
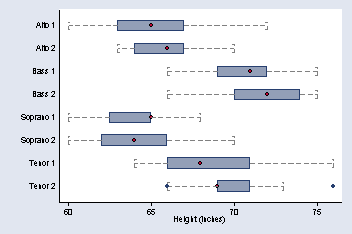
page 27, Figure 2.8
graph hbox height, over(voice) capsize(20) medmarker(msymbol(O)) ///
ytitle("Height (inches)") medtype(marker) ylabel( , nogrid) ///
cwhiskers lines(lpattern(dash))
To order the bars on the graph in the same way that they are ordered in the text, you need to create a new variable with values that correspond to the desired order.
gen v = 1
replace v = 2 if voice_part == "Soprano 2"
(30 real changes made)
replace v = 3 if voice_part == "Alto 1"
(35 real changes made)
replace v = 4 if voice_part == "Alto 2"
(27 real changes made)
replace v = 5 if voice_part == "Tenor 1"
(21 real changes made)
replace v = 6 if voice_part == "Tenor 2"
(21 real changes made)
replace v = 7 if voice_part == "Bass 1"
(39 real changes made)
replace v = 8 if voice_part == "Bass 2"
(26 real changes made)
label define v1 1 "Soprano 1" 2 "Soprano 2" 3 "Alto 1" 4 "Alto 2" ///
5 "Tenor 1" 6 "Tenor 2" 7 "Bass 1" 8 "Bass 2"
label values v v1
graph hbox height, over(v) capsize(20) medmarker(msymbol(O)) ///
ytitle("Height (inches)") medtype(marker) ylabel( , nogrid) cwhiskers ///
lines(lpattern(dash)) marker(1, msymbol(Oh))
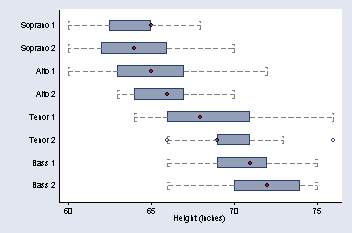
page 28, Figure 2.9
quantile height if voice_part == "Alto 1", xtitle("f-value") ///
ytitle("Alto 1 Height (inches)") xlabel( , grid) ylabel(60(4)72, ///
angle(0)) msymbol(Oh)
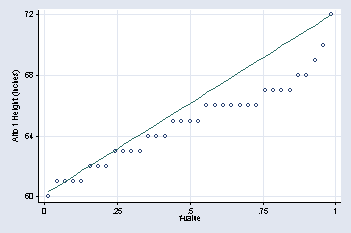
page 29, Figure 2.10
This has been skipped for now.
page 32, Figure 2.11
quantile height if voice_part == "Soprano 2", title(Soprano 2) ///
ylabel(60(5)75, angle(0)) xlabel(, grid) msymbol(Oh) xtitle("") ///
ytitle("") name(a, replace)
quantile height if voice_part == "Soprano 1", title(Soprano 1) ///
ylabel(60(5)75, angle(0)) xlabel(, grid) msymbol(Oh) xtitle("") ///
ytitle("") name(b, replace)
quantile height if voice_part == "Alto 1", title(Alto 1) ///
ylabel(60(5)75, angle(0)) xlabel(, grid) msymbol(Oh) xtitle("") ///
ytitle("") name(c, replace)
quantile height if voice_part == "Alto 2", title(Alto 2) ///
ylabel(60(5)75, angle(0)) xlabel(, grid) msymbol(Oh) xtitle("") ///
ytitle("") name(d, replace)
quantile height if voice_part == "Tenor 2", title(Tenor 2) ///
ylabel(60(5)75, angle(0)) xlabel(, grid) msymbol(Oh) xtitle("") ///
ytitle("") name(e, replace)
quantile height if voice_part == "Tenor 1", title(Tenor 1) ///
ylabel(60(5)75, angle(0)) xlabel(, grid) msymbol(Oh) xtitle("") ///
ytitle("") name(f, replace)
quantile height if voice_part == "Bass 2", title(Bass 2) ///
ylabel(60(5)75, angle(0)) xlabel(, grid) msymbol(Oh) xtitle("") ///
ytitle("") name(g, replace)
quantile height if voice_part == "Bass 1", title(Bass 1) ///
ylabel(60(5)75, angle(0)) xlabel(, grid) msymbol(Oh) xtitle("") ///
ytitle("") name(h, replace)
graph combine a b c d e f g h, l1title(Height (inches)) ///
cols(2) ysize(10)
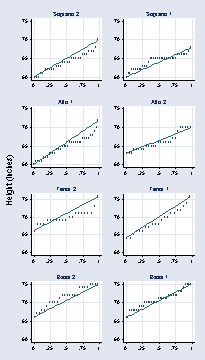
page 34, Figure 2.12
use "c:vizdatasinger.dta", clear gen v = 1 replace v = 2 if voice_part == "Soprano 2" (30 real changes made) replace v = 3 if voice_part == "Alto 1" (35 real changes made) replace v = 4 if voice_part == "Alto 2" (27 real changes made) replace v = 5 if voice_part == "Tenor 1" (21 real changes made) replace v = 6 if voice_part == "Tenor 2" (21 real changes made) replace v = 7 if voice_part == "Bass 1" (39 real changes made) replace v = 8 if voice_part == "Bass 2" (26 real changes made) label define v1 1 "Soprano 1" 2 "Soprano 2" 3 "Alto 1" 4 "Alto 2" /// 5 "Tenor 1" 6 "Tenor 2" 7 "Bass 1" 8 "Bass 2" label values v v1
NOTE: The ytick option is included so that the first dot is not right on the y-axis.
NOTE: The exclude0 option is necessary so that Stata does not start the x-axis at 0.
graph dot height, over(v) ytitle(Mean Height (inches)) exclude0 /// ylabel(60(2)70) ylabel(64(2)70) ymtick(65(2)71) ytick(63, tstyle(none))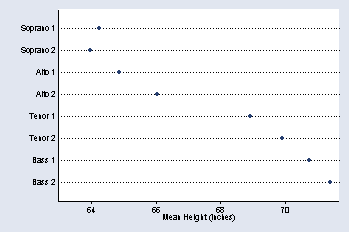
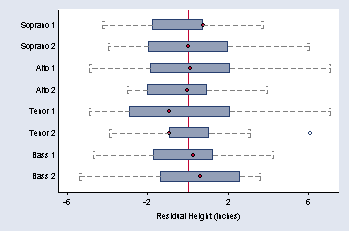
page 35, Figure 2.13
sort v
by v: egen mheight = mean(height)
by v: gen rheight = height - mheight
graph hbox rheight, over(v) medmarker(msymbol(O)) medtype(marker) ///
ytitle(" " "Residual Height (inches)") ylabel(-6 -2 2 6, nogrid) ///
yline(0) capsize(20) cwhiskers lines(lpattern(dash)) marker(1, msymbol(Oh))
page 37, Figure 2.14
gen r1 = height - mheight if voice_part == "Soprano 2"
(205 missing values generated)
gen r2 = height - mheight if voice_part == "Soprano 1"
(199 missing values generated)
gen r3 = height - mheight if voice_part == "Alto 2"
(208 missing values generated)
gen r4 = height - mheight if voice_part == "Alto 1"
(200 missing values generated)
gen r5 = height - mheight if voice_part == "Tenor 2"
(214 missing values generated)
gen r6 = height - mheight if voice_part == "Tenor 1"
(214 missing values generated)
gen r7 = height - mheight if voice_part == "Bass 2"
(209 missing values generated)
gen r8 = height - mheight if voice_part == "Bass 1"
(196 missing values generated)
qqplot r1 rheight, xtitle(" ") ytitle(" ") xlabel(-4 0 4, grid) ///
title(Soprano 2) ylabel(-4 0 4, angle(0)) xmtick(-2 2 6, grid) ///
ymtick(-2 2 6, grid) name(a, replace) msymbol(Oh)
qqplot r2 rheight, xtitle(" ") ytitle(" ") xlabel(-4 0 4, grid) ///
title(Soprano 1) ylabel(-4 0 4, angle(0)) xmtick(-2 2 6, grid) ///
ymtick(-2 2 6, grid) name(b, replace) msymbol(Oh)
qqplot r3 rheight, xtitle(" ") ytitle(" ") xlabel(-4 0 4, grid) ///
title(Alto 2) ylabel(-4 0 4, angle(0)) xmtick(-2 2 6, grid) ///
ymtick(-2 2 6, grid) name(c, replace) msymbol(Oh)
qqplot r4 rheight, xtitle(" ") ytitle(" ") xlabel(-4 0 4, grid) ///
title(Alto 1) ylabel(-4 0 4, angle(0)) xmtick(-2 2 6, grid) ///
ymtick(-2 2 6, grid) name(d, replace) msymbol(Oh)
qqplot r5 rheight, xtitle(" ") ytitle(" ") xlabel(-4 0 4, grid) ///
title(Tenor 2) ylabel(-4 0 4, angle(0)) xmtick(-2 2 6, grid) ///
ymtick(-2 2 6, grid) name(e, replace) msymbol(Oh)
qqplot r6 rheight, xtitle(" ") ytitle(" ") xlabel(-4 0 4, grid) ///
title(Tenor 1) ylabel(-4 0 4, angle(0)) xmtick(-2 2 6, grid) ///
ymtick(-2 2 6, grid) name(f, replace) msymbol(Oh)
qqplot r7 rheight, xtitle(" ") ytitle(" ") xlabel(-4 0 4, grid) ///
title(Bass 2) ylabel(-4 0 4, angle(0)) xmtick(-2 2 6, grid) ///
ymtick(-2 2 6, grid) name(g, replace) msymbol(Oh)
qqplot r8 rheight, xtitle(" ") ytitle(" ") xlabel(-4 0 4, grid) ///
title(Bass 1) ylabel(-4 0 4, angle(0)) xmtick(-2 2 6, grid) ///
ymtick(-2 2 6, grid) name(h, replace) msymbol(Oh)
graph combine a b c d e f g h, l1title(Residual Height (inches)) ///
b1title(Residual Height (inches)) cols(2) xsize(2)
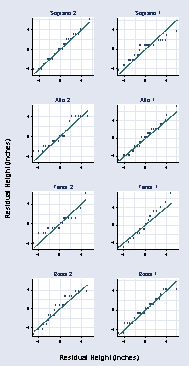
page 38, Figure 2.15
quantile rheight, ytitle(Residual Hight (inches)) /// xlabel(0 (.25)1) ylabel(-6(4)6, nogrid angle(0)) /// ymtick(-4 0 4) msymbol(Oh) xtitle(f-value)
page 39, Figure 2.16
qnorm rheight, ytitle(Residual Height (inches)) /// ylabel(-6(4)6) ymtick(-4 0 4) xmtick(-2 0 2) msymbol(Oh)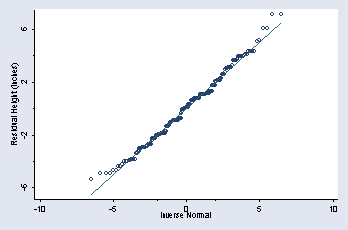
page 41, Figure 2.17
* panel 1
NOTE: We use the summ command to get the value for the mean of mheight, which we need to calculate the fitted values. This value is held in r(mean), which we use in the gen command. We do this so that we don’t have to know what the actual mean of mheight is.
summ mheight
Variable | Obs Mean Std. Dev. Min Max
-------------+--------------------------------------------------------
mheight | 235 67.29787 2.924444 63.96667 71.38461
gen x = mheight - r(mean)
quantile x, name(a, replace) xlabel(0 .5 1, grid) ylabel(-6(4)6, ///
angle(0)) xmtick(.25 .75, grid) ymtick(-4(4)4, grid) xtitle(" ") ///
ytitle(" ") title(Fitted Values)
* panel 2
quantile rheight, xlabel(0 .5 1, grid) ylabel(-6(4)6, angle(0)) ///
ymtick(-4 0 4, grid) xmtick(.25 .75, grid) msymbol(Oh) ///
title(Residuals) name(b, replace) xtitle(" ") ytitle(" ")
graph combine a b, b1title(f-value) l1title(Height (inches))
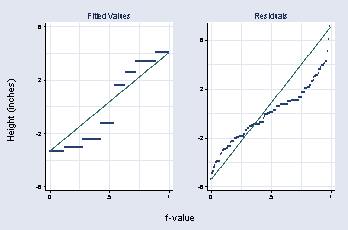
page 44, Figure 2.19
use "c:vizdatafusion.dta", clear
* panel 1
quantile time if nv_vv == "NV", msymbol(Oh) ylabel(0(10)40, grid ///
angle(0)) xlabel(0 .5 1, grid) xmtick(.25 .75, grid) name(a, replace) ///
xtitle(" ") ytitle(" ") title(NV)
* panel 2
quantile time if nv_vv == "VV", msymbol(Oh) ylabel(0(10)40, grid ///
angle(0)) xlabel(0 .5 1, grid) xmtick(.25 .75, grid) name(b, replace) ///
xtitle(" ") ytitle(" ") title(VV)
graph combine a b, b1title(f-value) l1title(Time (seconds))
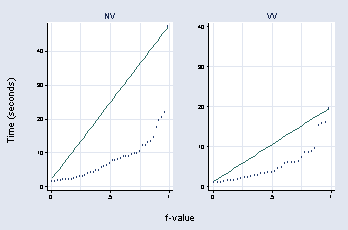
page 46, Figure 2.22
* panel 1
qnorm time if nv_vv == "NV", xlabel(, grid) ///
ytitle(" ") ylabel(0(10)40, grid) name(a, replace) title(NV) msymbol(Oh)
* panel 2
qnorm time if nv_vv == "VV", xlabel(, grid) ///
ytitle(" ") ylabel(0(10)40, grid) title(VV) msymbol(Oh) name(b, replace)
graph combine a b, l1title(Time (seconds))
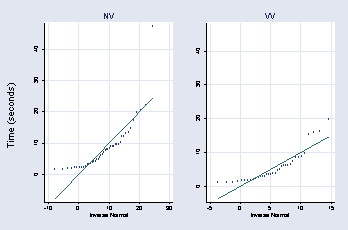
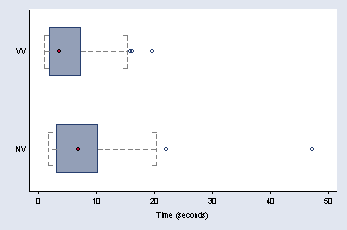
page 47, Figure 2.23
graph hbox time, over(nv_vv, descending) capsize(10) ///
medmarker(msymbol(O)) ytitle(" " "Time (seconds)") medtype(marker) ///
ylabel(, nogrid) cwhiskers lines(lpattern(dash)) marker(1, msymbol(Oh))
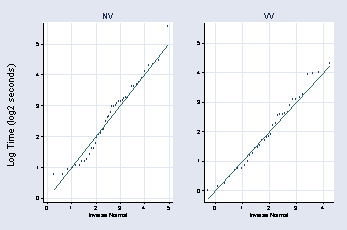
page 49, Figure 2.24
* panel 1
gen log2time = ln(time)/ln(2)
qnorm log2time if nv_vv == "NV", title(NV) msymbol(Oh) ///
ytitle(" ") ylabel(0(1)5, grid angle(0)) xlabel(, grid) name(a, replace)
* panel 2
qnorm log2time if nv_vv == "VV", title("VV") name(b, replace) msymbol(Oh) ///
ytitle(" ") ylabel(0(1)5, grid angle(0)) xlabel(, grid)
graph combine a b, l1title(Log Time (log2 seconds))
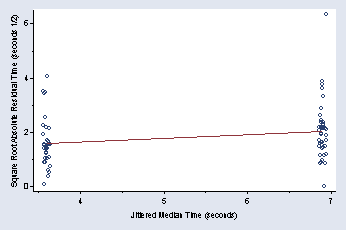
page 50, Figure 2.25
encode nv_vv, gen(vv)
qreg time vv, q(50)
Iteration 1: WLS sum of weighted deviations = 351.70419
Iteration 1: sum of abs. weighted deviations = 352.70001
Iteration 2: sum of abs. weighted deviations = 341.60001
Iteration 3: sum of abs. weighted deviations = 337.29999
Median regression Number of obs = 78
Raw sum of deviations 350 (about 4.9000001)
Min sum of deviations 337.3 Pseudo R2 = 0.0363
------------------------------------------------------------------------------
time | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
vv | -3.3 1.622147 -2.03 0.045 -6.530785 -.0692153
_cons | 10.2 2.486812 4.10 0.000 5.247085 15.15292
------------------------------------------------------------------------------
predict r, res
gen r2 = sqrt(abs(r))
predict p
(option xb assumed; fitted values)
graph twoway (scatter r2 p, jitter(3) msymbol(Oh)) (lfit r2 p), ///
xtitle(" " "Jittered Median Time (seconds)") xlabel(4(1)7) ///
ytitle("Square Root Absolute Residual Time (seconds 1/2)") ///
ylabel(0(2)6, nogrid angle(0)) ymtick(1 3 5) legend(off) xmtick(3.5(1)6.5)
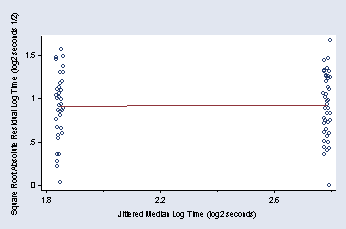
page 51, Figure 2.26
qreg log2time vv, q(50)
Iteration 1: WLS sum of weighted deviations = 76.575202
Iteration 1: sum of abs. weighted deviations = 76.390258
Iteration 2: sum of abs. weighted deviations = 76.119386
Iteration 3: sum of abs. weighted deviations = 76.041383
Median regression Number of obs = 78
Raw sum of deviations 79.63833 (about 2.2927818)
Min sum of deviations 76.04138 Pseudo R2 = 0.0452
------------------------------------------------------------------------------
log2time | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
vv | -.9385995 .4082099 -2.30 0.024 -1.75162 -.1255789
_cons | 3.725196 .6258012 5.95 0.000 2.478805 4.971587
------------------------------------------------------------------------------
predict r51, res
gen r512 = sqrt(abs(r51))
predict p51
(option xb assumed; fitted values)
graph twoway (scatter r512 p51, jitter(3) msymbol(Oh)) ///
(lfit r512 p51), xtitle(" " "Jittered Median Log Time (log2 seconds)") ///
xlabel(1.8 2.2 2.6) xmtick(2 2.4 2.8) ///
ytitle(Square Root Absolute Residual Log Time (log2 seconds 1/2)) ///
ylabel(0(.5)1.5, nogrid angle(0)) legend(off) graphregion(margin(t=15))
page 52, Figure 2.27
gen vv1 = time if vv == 1
(35 missing values generated)
gen vv2 = time if vv == 2
(43 missing values generated)
qqplot vv2 vv1, xtitle(" " "NV Time (seconds)") ytitle(VV Time ///
(seconds)) ymtick(5(10)45) xmtick(5(10)45) ylabel( ,nogrid angle(0)) ///
title("") msymbol(Oh)
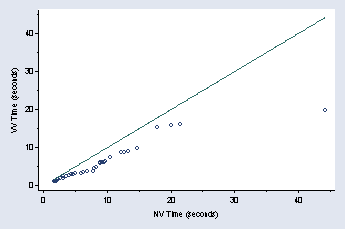
page 53, Figure 2.28
gen lognv = log(time) / log(2) if vv == 1
(35 missing values generated)
gen logvv = log(time) / log(2) if vv == 2
(43 missing values generated)
qqplot logvv lognv, xtitle(" " "Log NV Time (log 2 seconds)") ///
ytitle("Log VV Time (log 2 seconds)") xlabel(0(1)5) ylabel(0(1)5) ///
ylabel( ,nogrid angle(0)) title("") msymbol(Oh)
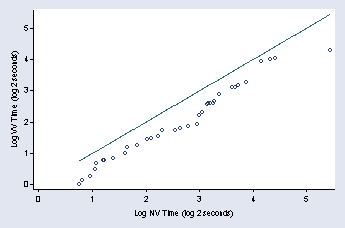
page 53, Figure 2.29
tmdplot logvv lognv, ytitle(Mean (log2 seconds)) xtitle(Difference ///
(log2 seconds)) msymbol(Oh) ylabel(-1(.2)-.2, nogrid angle(0)) ///
ymtick(-1.1(.1)-.3) xlabel(1(1)5) xmtick(.5(1)4.5) title("")
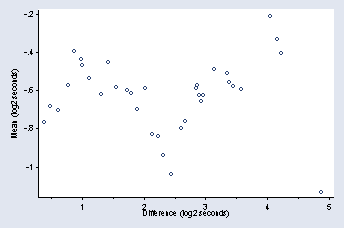
page 54, Figure 2.30
xi: regress log2time i.vv
i.vv _Ivv_1-2 (naturally coded; _Ivv_1 omitted)
Source | SS df MS Number of obs = 78
-------------+------------------------------ F( 1, 76) = 5.38
Model | 7.44477409 1 7.44477409 Prob > F = 0.0231
Residual | 105.212331 76 1.38437277 R-squared = 0.0661
-------------+------------------------------ Adj R-squared = 0.0538
Total | 112.657105 77 1.46307928 Root MSE = 1.1766
------------------------------------------------------------------------------
log2time | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
_Ivv_2 | -.6211619 .2678586 -2.32 0.023 -1.154649 -.0876753
_cons | 2.625721 .1794289 14.63 0.000 2.268357 2.983084
------------------------------------------------------------------------------
predict rpooled, resid
regress log2time vv if vv==1
Source | SS df MS Number of obs = 43
-------------+------------------------------ F( 0, 42) = 0.00
Model | 0 0 . Prob > F = .
Residual | 57.8833874 42 1.37817589 R-squared = 0.0000
-------------+------------------------------ Adj R-squared = 0.0000
Total | 57.8833874 42 1.37817589 Root MSE = 1.174
------------------------------------------------------------------------------
log2time | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
vv | (dropped)
_cons | 2.625721 .1790268 14.67 0.000 2.26443 2.987011
------------------------------------------------------------------------------
predict rvv1 if e(sample), resid
(35 missing values generated)
regress log2time vv if vv==2
Source | SS df MS Number of obs = 35
-------------+------------------------------ F( 0, 34) = 0.00
Model | 0 0 . Prob > F = .
Residual | 47.3289433 34 1.39202774 R-squared = 0.0000
-------------+------------------------------ Adj R-squared = 0.0000
Total | 47.3289433 34 1.39202774 Root MSE = 1.1798
------------------------------------------------------------------------------
log2time | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
vv | (dropped)
_cons | 2.004559 .1994297 10.05 0.000 1.599269 2.409849
------------------------------------------------------------------------------
predict rvv2 if e(sample), resid
(43 missing values generated)
qqplot rvv1 rpooled, xlabel(-2(1)2, grid) ylabel(-2(1)3, grid angle(0)) ///
xtitle("") ytitle("") title("NV") name(rvv1, replace) msymbol(Oh)
qqplot rvv2 rpooled, xlabel(-2(1)2, grid) ylabel(-2(1)3, grid angle(0)) ///
xtitle("") ytitle("") title("VV") name(rvv2, replace) msymbol(Oh)
graph combine rvv1 rvv2, b1title("Residual Log Time (log2 seconds)") ///
l1title(Residual Log Time (log2 seconds))
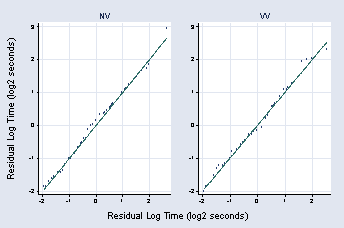
page 55, Figure 2.31
qnorm rpooled, ylabel(-2(1)3, angle(0)) /// ytitle(Residual Log Time (log2 seconds)) msymbol(Oh)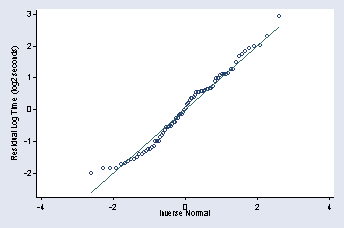
page 55, Figure 2.32
xi: regress log2time i.vv
i.vv _Ivv_1-2 (naturally coded; _Ivv_1 omitted)
Source | SS df MS Number of obs = 78
-------------+------------------------------ F( 1, 76) = 5.38
Model | 7.44477409 1 7.44477409 Prob > F = 0.0231
Residual | 105.212331 76 1.38437277 R-squared = 0.0661
-------------+------------------------------ Adj R-squared = 0.0538
Total | 112.657105 77 1.46307928 Root MSE = 1.1766
------------------------------------------------------------------------------
log2time | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
_Ivv_2 | -.6211619 .2678586 -2.32 0.023 -1.154649 -.0876753
_cons | 2.625721 .1794289 14.63 0.000 2.268357 2.983084
------------------------------------------------------------------------------
predict fit
(option xb assumed; fitted values)
summ fit
Variable | Obs Mean Std. Dev. Min Max
-------------+--------------------------------------------------------
fit | 78 2.346994 .3109427 2.004559 2.625721
gen fitted = fit - 2.346994
quantile fitted, ylabel(-2(1)3) xlabel(0 .5 1, grid) msymbol(Oh) ///
name(fit, replace) xtitle("") ytitle(Log Time (log2 seconds))
quantile rpooled, name(res, replace) xlabel(0 .5 1, grid) xlabel("") ///
xtitle("") ytitle("") msymbol(Oh)
graph combine fit res, b1title(f-value)
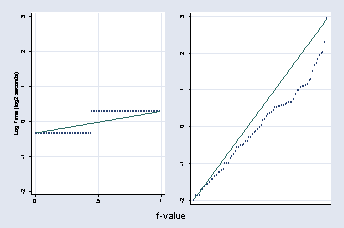
page 57, Figure 2.33
* panel 1
qnorm time if nv_vv == "VV", ylabel(0 10 20) name(a, replace) title(1) ///
ytitle("") xtitle("") xlabel( , grid) msymbol(Oh)
* panel 2
gen time2 = (time)^.25
qnorm time2 if nv_vv == "VV", ylabel(1 1.5 2) name(b, replace) ///
title(0.25) ytitle("") xtitle("") xlabel( , grid) msymbol(Oh)
* panel 3
gen time3 = (time)^-.25
qnorm time3 if nv_vv == "VV", ylabel(.5 .75 1) name(c, replace) ///
title(-0.25) ytitle("") xtitle("") xlabel( , grid) msymbol(Oh)
* panel 4
gen time4 = (time)^-1
qnorm time4 if nv_vv == "VV", ylabel(0 .5 1) name(d, replace) ///
ylabel(-2 0 2) title(-1) ytitle("") xtitle("") xlabel( , grid) msymbol(Oh)
* panel 5
gen time5 = (time)^.5
qnorm time5 if nv_vv == "VV", ylabel(1(1)4) name(e, replace) ///
title(.5) ytitle("") xtitle("") xlabel( , grid) msymbol(Oh)
* panel 6
gen time6 = ln(time)
qnorm time6 if nv_vv == "VV", ylabel(0(1)3) name(f, replace) ///
title(0) ytitle("") xtitle("") xlabel( , grid) msymbol(Oh)
* panel 7
gen time7 = (time)^-.5
qnorm time7 if nv_vv == "VV", ylabel(.2 .6 1) name(g, replace) ///
title(-0.5) ytitle("") xtitle("") xlabel( , grid) msymbol(Oh)
graph combine a b e c f d g, cols(2) rows(4) l1title("VV Time") ///
ysize(5) xsize(3) holes(2)
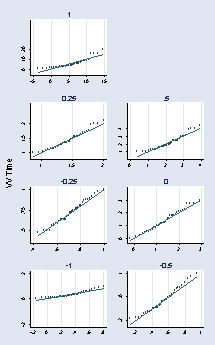
page 59, Figure 2.34
use "c:vizdatafoodweb.dta", clear
* panel 1
quantile mean_length if dimension == "Three", ylabel(2(1)6, grid ///
angle(0)) xmtick(.2 .75, grid) xlabel(0 .5 1, grid) name(a, replace) ///
ytitle("") xtitle("") title("Three") msymbol(Oh)
* panel 2
quantile mean_length if dimension == "Mixed", ylabel(2(1)6, grid ///
angle(0)) xmtick(.2 .75, grid) xlabel(0 .5 1, grid) name(b, replace) ///
ytitle("") xtitle("") title("Mixed") msymbol(Oh)
* panel 3
quantile mean_length if dimension == "Two", ylabel(2(1)6, grid angle(0)) ///
msymbol(Oh) xmtick(.2 .75, grid) xlabel(0 .5 1, grid) name(c, replace) ///
ytitle("") xtitle("") xmtick(.25 .75) title("Two")
graph combine a b c, cols(1) b1title("f-value") ///
l1title("Chain Length") xsize(3) ysize(5)
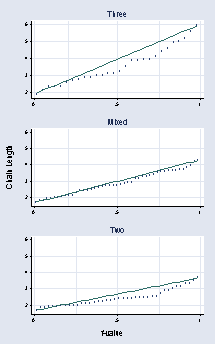
page 60, Figure 2.35
encode dimension, gen(dim) sort dim by dim: egen mcl = median(mean_length) by dim: gen absr = sqrt(abs(mean_length - mcl)) by dim: egen mr = median(absr) sort mcl graph twoway (scatter absr mcl, jitter(5) msymbol(Oh)) /// (scatter mr mcl, c(L)), ylabel(0(.5)1.5, angle(0) nogrid) /// xtitle(Jittered Median Chain Length) ytitle(Square Root Absolute /// Residual Chain Length) legend(off)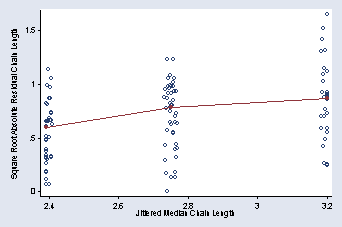
page 61, Figure 2.36
* panel 1
qnorm mean_length if dimension == "Three", ylabel(2(1)6, grid ///
angle(0)) title("Three") name(a, replace) ytitle("") xlabel( ,grid) ///
xtitle("") msymbol(Oh)
* panel 2
qnorm mean_length if dimension == "Mixed", ylabel(2(1)6, grid ///
angle(0)) title("Mixed") name(b, replace) ytitle("") xlabel( ,grid) ///
xtitle("") msymbol(Oh)
* panel 3
qnorm mean_length if dimension == "Two", ylabel(2(1)6, grid angle(0)) ///
title("Two") name(c, replace) ytitle("") xlabel( ,grid) xtitle("") ///
msymbol(Oh)
graph combine a b c, l1title("Chain Length") cols(1) xsize(3) ysize(6)
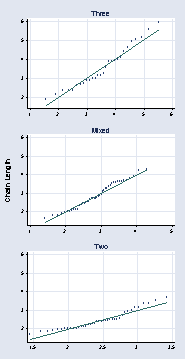
page 62, Figure 2.37
gen log2cl = ln(mean_length) / ln(2)
sort dim
by dim: egen mll = median(log2cl )
by dim: gen absres = sqrt(abs(log2cl - mll))
by dim: egen mres = median(absres)
sort mll
graph twoway (scatter absres mll, jitter(5) msymbol(Oh)) ///
(scatter mres mll, c(L)), ///
xtitle(" " "Jittered Median Log2 Chain Length") ///
xlabel(1.3(.1)1.7) xmtick(1.25(.1)1.65) ///
ytitle(Square Root Absolute Residual Log2 Chain Length) ///
ylabel(0(.2).8, nogrid) ymtick(.1(.1).9) legend(off)
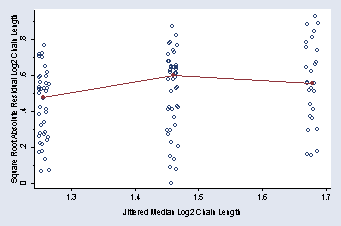
page 63, Figure 2.38
* panel 1
qnorm log2cl if dimension == "Three", ylabel(1(.5)2.5, grid angle(0)) ///
title("Three") name(a, replace) ytitle("") xlabel( ,grid) xtitle("") ///
msymbol(Oh)
* panel 2
qnorm log2cl if dimension == "Mixed", ylabel(1(.5)2.5, grid angle(0)) ///
title("Mixed") name(b, replace) ytitle("") xlabel( ,grid) xtitle("") ///
msymbol(Oh)
* panel 3
qnorm log2cl if dimension == "Two", ylabel(1(.5)2.5, grid angle(0)) ///
title("Two") name(c, replace) ytitle("") xlabel( ,grid) xtitle("") ///
msymbol(Oh)
graph combine a b c, l1title("Chain Length") cols(1) xsize(3) ysize(6)
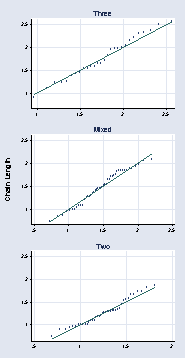
page 64, Figure 2.39
gen invl = 1/mean_length
sort dim
by dim: egen minv = median(invl)
by dim: gen absresinv = sqrt(abs(invl - minv))
by dim: egen mresinv = median(absresinv)
sort minv
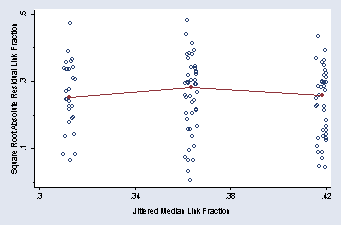
graph twoway (scatter absresinv minv, jitter(5) msymbol(Oh)) ///
(scatter mresinv minv, c(L)), xtitle(" " "Jittered Median Link Fraction") ///
xlabel(.3(.04).42) ytitle(Square Root Absolute Residual Link Fraction) ///
ylabel(.1(.2).5, nogrid) ymtick(0(.2).4) legend(off)
page 65, Figure 2.40
* panel 1
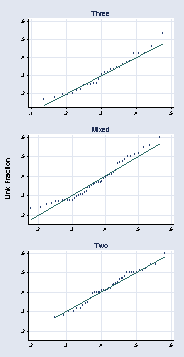
qnorm invl if dimension == "Three", ylabel(.2(.1).6, grid angle(0)) ///
title("Three") name(a, replace) ytitle("") xlabel( ,grid) xtitle("") ///
msymbol(Oh)
* panel 2
qnorm invl if dimension == "Mixed", ylabel(.2(.1).6, grid angle(0)) ///
title("Mixed") name(b, replace) ytitle("") xlabel( ,grid) xtitle("") ///
msymbol(Oh)
* panel 3
qnorm invl if dimension == "Two", ylabel(.2(.1).6, grid angle(0)) ///
title("Two") name(c, replace) ytitle("") xlabel( ,grid) xtitle("") ///
msymbol(Oh)
graph combine a b c, l1title(Link Fraction) cols(1) xsize(3) ysize(6)
page 66, Figure 2.41
sort dim
by dim: egen meanl = mean(invl)
by dim: gen resid = invl - meanl
gen r2 = resid if dim == 3
(73 missing values generated)
gen r2e = resid if dim ~=3
(40 missing values generated)
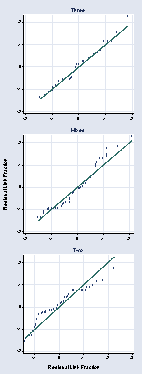
qqplot r2 r2e, title(Two) name(a, replace) msymbol(Oh) ///
ylabel(, angle(0)) xlabel(, grid) xmtick(-.05 .05 .15) ///
ymtick(-.05 .05 .15) xtitle("") ytitle("")
gen rm = resid if dim == 1
(68 missing values generated)
gen rme = resid if dim ~=1
(45 missing values generated)
qqplot rm rme, title(Mixed) name(b, replace) msymbol(Oh) ///
ylabel(, angle(0)) xlabel(, grid) xmtick(-.05 .05 .15) ///
ymtick(-.05 .05 .15) xtitle("") ytitle("")
gen r3 = resid if dim == 2
(85 missing values generated)
gen r3e = resid if dim ~=2
(28 missing values generated)
qqplot r3 r3e, title(Three) name(c, replace) msymbol(Oh) ///
ylabel(, angle(0)) xlabel(, grid) xmtick(-.05 .05 .15) ///
ymtick(-.05 .05 .15) xtitle("") ytitle("")
graph combine c b a, b1title(Residual Link Fraction) ///
l1title(Residual Link Fraction) cols(1) xsize(1.5)
page 67, Figure 2.42
* panel 1
sort dim
by dim: egen mml = mean(invl)
summ mml
Variable | Obs Mean Std. Dev. Min Max
-------------+--------------------------------------------------------
mml | 113 .3749383 .045431 .3075056 .4257752
gen xx = mml - r(mean)
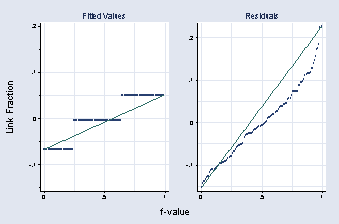
quantile xx, msymbol(Oh) name(a, replace) xlabel(0 .5 1, grid) ///
ylabel(-.1(.1).2, angle(0)) xtitle("") ytitle("") title(Fitted Values) ///
xmtick(.25 .75, grid) ymtick(-.15(.1).15, grid)
* panel 2
quantile resid, title(Residuals) name(b, replace) msymbol(Oh) ///
xlabel(0 .5 1, grid) xtitle("") ylabel(-.1(.1).2, angle(0)) ytitle("") ///
xmtick(.25 .75, grid) ymtick(-.15(.1).15, grid)
graph combine a b, b1title( f-value) l1title(Link Fraction)
page 69, Figure 2.43
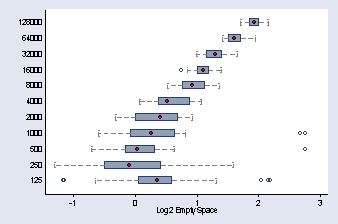
use "c:vizdatabin.dta", clear
gen log2es = ln(empty_space) / ln(2)
graph hbox log2es, over(number_runs, descending) ylabel( , nogrid) ///
capsize(20) medmarker(msymbol(O)) ytitle("Log2 Empty Space") ///
medtype(marker) cwhiskers lines(lpattern(dash)) marker(1, msymbol(Oh))
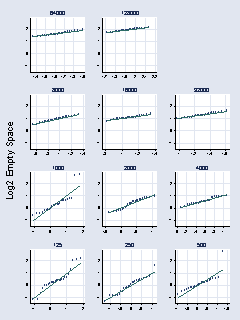
page 71, Figure 2.44
* panel 1
qnorm log2es if number_runs == 64000, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(a, replace) title(64000) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 2
qnorm log2es if number_runs == 8000, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(b, replace) title(8000) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 3
qnorm log2es if number_runs == 1000, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(c, replace) title(1000) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 4
qnorm log2es if number_runs == 125, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(d, replace) title(125) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 5
qnorm log2es if number_runs == 128000, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(e, replace) title(128000) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 6
qnorm log2es if number_runs == 16000, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(f, replace) title(16000) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 7
qnorm log2es if number_runs == 2000, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(g, replace) title(2000) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 8
qnorm log2es if number_runs == 250, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(h, replace) title(250) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 9
qnorm log2es if number_runs == 32000, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(i, replace) title(32000) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 10
qnorm log2es if number_runs == 4000, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(j, replace) title(4000) ytitle("") xtitle("") ///
msymbol(Oh)
* panel 11
qnorm log2es if number_runs == 500, ylabel(-1(1)2, grid angle(0)) ///
xlabel( , grid) name(k, replace) title(500) ytitle("") xtitle("") ///
msymbol(Oh)
graph combine a e b f i c g j d h k, cols(3) rows(4) ///
l1title(Log2 Empty Space) xsize(5) ysize(7) holes(3)
page 74, Figure 2.45
NOTE: You can use either qreg or egen to compute the necessary values.
* qreg log2es number_runs * predict yhat egen yhat = median(log2es), by(number_runs) gen e = log2es - yhat gen abse = abs(e) egen mad = median(abse), by(number_runs) gen ssr45 = e/mad qnorm ssr45 if number_runs == 64000, ylabel(-5(5)10, grid angle(0)) /// name(a, replace) title(64000) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 128000, ylabel(-5(5)10, grid angle(0)) /// name(b, replace) title(128000) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 8000, ylabel(-5(5)10, grid angle(0)) /// name(c, replace) title(8000) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 16000, ylabel(-5(5)10, grid angle(0)) /// name(d, replace) title(16000) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 32000, ylabel(-5(5)10, grid angle(0)) /// name(e, replace) title(32000) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 1000, ylabel(-5(5)10, grid angle(0)) /// name(f, replace) title(1000) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 2000, ylabel(-5(5)10, grid angle(0)) /// name(g, replace) title(2000) xlabel(, grid) /// xmtick(-2.5 2.5, grid tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 4000, ylabel(-5(5)10, grid angle(0)) /// name(h, replace) title(4000) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 125, ylabel(-5(5)10, grid angle(0)) /// name(i, replace) title(125) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 250, ylabel(-5(5)10, grid angle(0)) /// name(j, replace) title(250) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 500, ylabel(-5(5)10, grid angle(0)) /// name(k, replace) title(500) xlabel(, grid) xmtick(-2.5 2.5, grid /// tstyle(none)) msymbol(Oh) graph combine a b c d e f g h i j k, l1title(Spread-Standardized /// Residual Log2 Empty Space) hole(3) rows(4) xsize(3) ysize(5)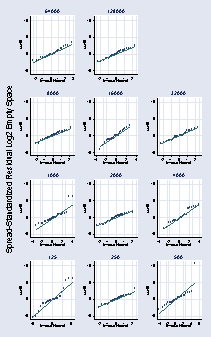
page 75, Figure 2.46
sort number_runs
by number_runs: egen sn = mad(log2es)
by number_runs: egen ln = median(log2es)
by number_runs: gen resid = (log2es- ln )/sn
gen r2000 = resid if number_runs == 2000
(250 missing values generated)
gen r2000e = resid if number_runs ~=2000
(25 missing values generated)
qqplot r2000 r2000e, name(f, replace) title(2000) xtitle("") ytitle("") ///
xlabel(, grid) msymbol(Oh) ylabel(-4 0 4, angle(0)) xlabel(-4 0 4) ///
xmtick(-2 2, grid) ymtick(-2 2, grid)
gen r4000 = resid if number_runs == 4000
(250 missing values generated)
gen r4000e = resid if number_runs ~=4000
(25 missing values generated)
qqplot r4000 r4000e, name(g, replace) title(4000) xtitle("") ytitle("") ///
xlabel(, grid) msymbol(Oh) ylabel(-4 0 4, angle(0)) xlabel(-4 0 4) ///
xmtick(-2 2, grid) ymtick(-2 2, grid)
gen r8000 = resid if number_runs == 8000
(250 missing values generated)
gen r8000e = resid if number_runs ~=8000
(25 missing values generated)
qqplot r8000 r8000e, name(d, replace) title(8000) xtitle("") ytitle("") ///
xlabel(, grid) msymbol(Oh) ylabel(-4 0 4, angle(0)) xlabel(-4 0 4) ///
xmtick(-2 2, grid) ymtick(-2 2, grid)
gen r16000 = resid if number_runs == 16000
(250 missing values generated)
gen r16000e = resid if number_runs ~=16000
(25 missing values generated)
qqplot r16000 r16000e, name(e, replace) title(16000) xtitle("") ///
ytitle("") xlabel(, grid) msymbol(Oh) ylabel(-4 0 4, angle(0)) ///
xlabel(-4 0 4) xmtick(-2 2, grid) ymtick(-2 2, grid)
gen r32000 = resid if number_runs == 32000
(250 missing values generated)
gen r32000e = resid if number_runs ~=32000
(25 missing values generated)
qqplot r32000 r32000e, name(b, replace) title(32000) xtitle("") ///
ytitle("") xlabel(, grid) msymbol(Oh) ylabel(-4 0 4, angle(0)) ///
xlabel(-4 0 4) xmtick(-2 2, grid) ymtick(-2 2, grid)
gen r64000 = resid if number_runs == 64000
(250 missing values generated)
gen r64000e = resid if number_runs ~=64000
(25 missing values generated)
qqplot r64000 r64000e, name(c, replace) title(64000) xtitle("") ///
ytitle("") xlabel(, grid) msymbol(Oh) ylabel(-4 0 4, angle(0)) ///
xlabel(-4 0 4) xmtick(-2 2, grid) ymtick(-2 2, grid)
gen r128000 = resid if number_runs == 128000
(250 missing values generated)
gen r128000e = resid if number_runs ~=128000
(25 missing values generated)
qqplot r128000 r128000e, name(a, replace) title(128000) xtitle("") ///
ytitle("") xlabel(, grid) msymbol(Oh) ylabel(-4 0 4, angle(0)) ///
xlabel(-4 0 4) xmtick(-2 2, grid) ymtick(-2 2, grid)
graph combine a b c d e f g, b1title(Spread-Standardized Residual ///
Log2 Empty Space) l1title(Spread-Standardized Residual Log2 Empty Space) ///
cols(2) holes(2) xsize(2)
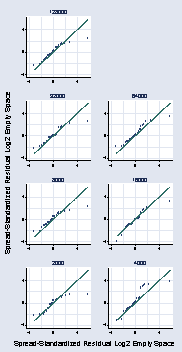
page 76, Figure 2.47
preserve drop if number_runs <=1000 (100 observations deleted) qnorm resid, msymbol(Oh) ytitle(Spread-Standardized Residual Log2 /// Empty Space) ylabel(-4(2)4, angle(0) nogrid)restore
page 77, Figure 2.48
gen log2nwts = ln(number_runs) / ln(2)
We have omitted the line.
graph twoway (scatter yhat log2nwts, msymbol(Oh)), ylabel(0(.5)2, /// angle(0)) xlabel(7(2)17) xtitle(Log2 Number of Weights) /// ytitle(Median Log2 Empty Space) legend(off)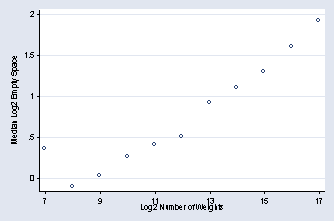
page 77, Figure 2.49
gen logn249 = log(number_runs)/log(2)
gen bin249 = log(empty_space)/log(2)
by number_runs: egen sn249 = mad(bin249)
gen logmad249 = log(sn249)/log(2)
graph scatter logmad249 logn249, msymbol(Oh) ylabel(, angle(0) nogrid) ///
ytitle(Log2 Mad of Log2 Empty Space) xtitle(" " "Log2 Number of Weights")
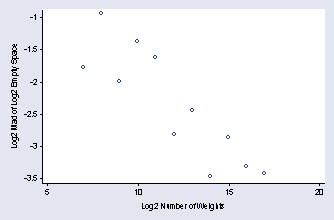
page 78, Figure 2.50
sort number_runs by number_runs: egen mad78 = mad(log2es) egen min78 = min(mad78) by number_runs: gen rs78 = mad78/min78 gen lnrs78 = ln(rs78)/ln(2) by number_runs: egen median78 = median(log2es) graph scatter lnrs78 median78, xtitle(Median Log2 Empty Space) /// ytitle(Log2 Relative Spread) ylabel(0(.5)2.5, nogrid angle(0)) /// xlabel(0(.5)2) msymbol(Oh)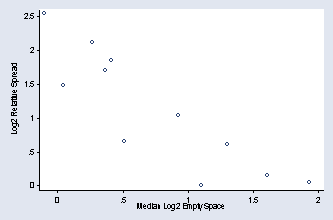
page 79, Figure 2.51
sort number_runs by number_runs: egen mad79 = mad(empty_space) egen min79 = min(mad79) by number_runs: gen rs79 = mad79/min79 gen lnrs79 = ln(rs79) by number_runs: egen median79 = median(empty_space) graph scatter lnrs79 median79, xtitle(Median Empty Space) /// ytitle(Log2 Relative Spread) ylabel(0(.4)1.2, nogrid angle(0)) /// xlabel(1(.7)3.8) msymbol(Oh) ymtick(.2(.4)1.4)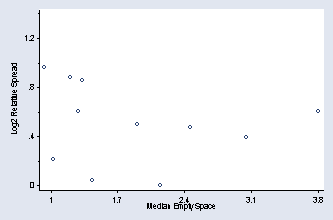
page 81, Figure 2.52
preserve drop if ssr45 > 4 | ssr45 < -4 (9 observations deleted) qnorm ssr45 if number_runs == 64000, ylabel(-4 0 4, grid angle(0)) /// name(a, replace) title(64000) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 128000, ylabel(-4 0 4, grid angle(0)) /// name(b, replace) title(128000) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 8000, ylabel(-4 0 4, grid angle(0)) /// name(c, replace) title(8000) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 16000, ylabel(-4 0 4, grid angle(0)) /// name(d, replace) title(16000) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 32000, ylabel(-4 0 4, grid angle(0)) /// name(e, replace) title(32000) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 1000, ylabel(-4 0 4, grid angle(0)) /// name(f, replace) title(1000) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 2000, ylabel(-4 0 4, grid angle(0)) /// name(g, replace) title(2000) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 4000, ylabel(-4 0 4, grid angle(0)) /// name(h, replace) title(4000) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 125, ylabel(-4 0 4, grid angle(0)) /// name(i, replace) title(125) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 250, ylabel(-4 0 4, grid angle(0)) /// name(j, replace) title(250) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) qnorm ssr45 if number_runs == 500, ylabel(-4 0 4, grid angle(0)) /// name(k, replace) title(500) xlabel(, grid) ymtick(-2 2, grid /// tstyle(none)) msymbol(Oh) graph combine a b c d e f g h i j k, l1title(Spread-Standardized /// Residual Log2 Empty Space) hole(3) rows(4) xsize(4) ysize(5)restore