This FAQ is an elaboration of a FAQ by Allen McDowell of StataCorp. and Nicholas J. Cox of Durham University. Please see www.stata.com/support/faqs/stat/logit.html for the original.
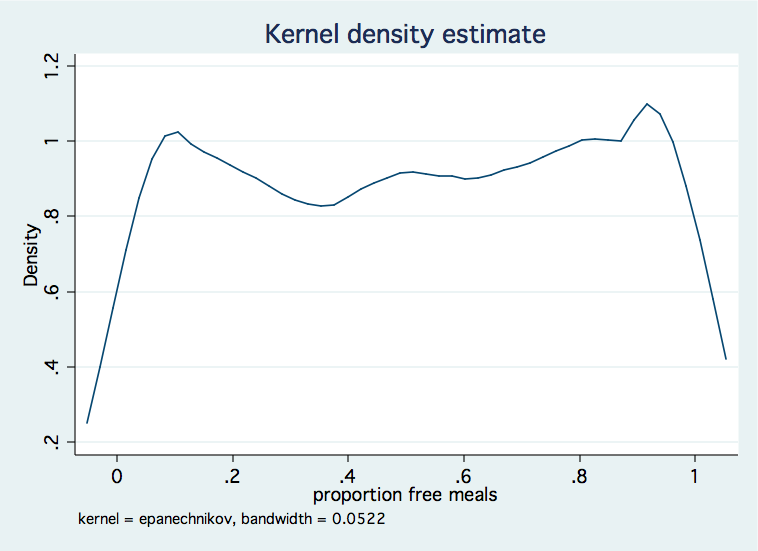
Proportion data has values that fall between zero and one. Naturally, it would be nice to have the predicted values also fall between zero and one. One way to accomplish this is to use a generalized linear model (glm) with a logit link and the binomial family. We will include the robust option in the glm model to obtain robust standard errors which will be particularly useful if we have misspecified the distribution family.
We will demonstrate this using a dataset in which the dependent variable, meals, is the proportion of students receiving free or reduced priced meals at school.
Next, we will compute predicted scores from the model and transform them back so that they are scaled the same way as the original proportions.use https://stats.idre.ucla.edu/stat/stata/faq/proportion, clear /* kernel density distribution of meals */ kdensity mealsglm meals yr_rnd parented api99, link(logit) family(binomial) robust nolog note: meals has non-integer values Generalized linear models No. of obs = 4257 Optimization : ML Residual df = 4253 Scale parameter = 1 Deviance = 395.8141242 (1/df) Deviance = .093067 Pearson = 374.7025759 (1/df) Pearson = .0881031 Variance function: V(u) = u*(1-u/1) [Binomial] Link function : g(u) = ln(u/(1-u)) [Logit] AIC = .7220973 Log pseudolikelihood = -1532.984106 BIC = -35143.61 ------------------------------------------------------------------------------ | Robust meals | Coef. Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------- yr_rnd | .0482527 .0321714 1.50 0.134 -.0148021 .1113074 parented | -.7662598 .0390715 -19.61 0.000 -.8428386 -.6896811 api99 | -.0073046 .0002156 -33.89 0.000 -.0077271 -.0068821 _cons | 6.75343 .0896767 75.31 0.000 6.577667 6.929193 ------------------------------------------------------------------------------
predict premeals1
(option mu assumed; predicted mean meals)
(164 missing values generated)
summarize meals premeals1 if e(sample)
Variable | Obs Mean Std. Dev. Min Max
-------------+--------------------------------------------------------
meals | 4257 .5165962 .3100389 0 1
premeals1 | 4257 .5165962 .2849672 .0220988 .9770855
As a contrast, let's run the same analysis without the transformation. We will then
graph the original dependent variable and the two predicted variables against api99.
regress meals yr_rnd parented api99
Source | SS df MS Number of obs = 4257
-------------+------------------------------ F( 3, 4253) = 6752.22
Model | 338.097096 3 112.699032 Prob > F = 0.0000
Residual | 70.985399 4253 .016690665 R-squared = 0.8265
-------------+------------------------------ Adj R-squared = 0.8264
Total | 409.082495 4256 .096119007 Root MSE = .12919
------------------------------------------------------------------------------
meals | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
yr_rnd | .0024454 .0054678 0.45 0.655 -.0082742 .013165
parented | -.1298907 .0048289 -26.90 0.000 -.1393579 -.1204234
api99 | -.0014118 .0000269 -52.40 0.000 -.0014646 -.0013589
_cons | 1.766162 .0134423 131.39 0.000 1.739808 1.792516
------------------------------------------------------------------------------
predict preols
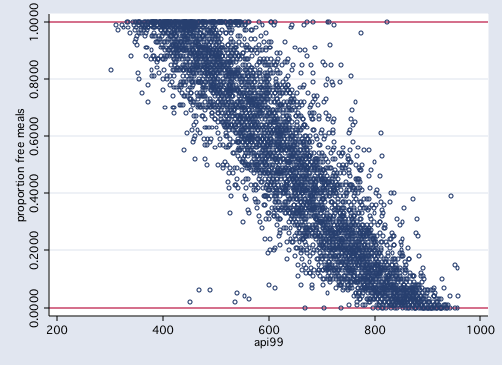
/* figure 1: proportion dependent variable */
graph twoway scatter meals api99, yline(0 1) msym(oh)
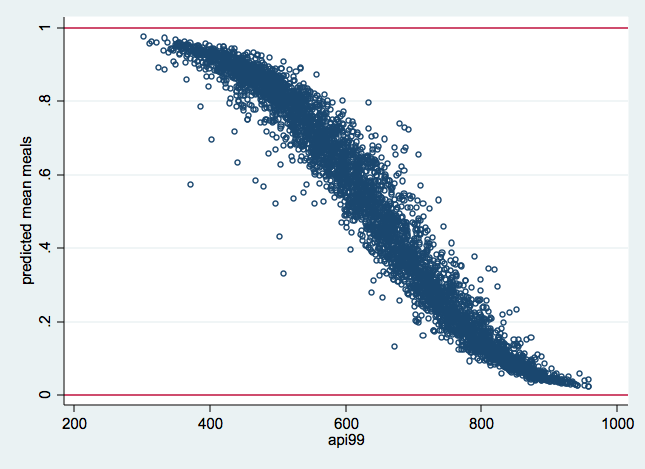
 /* figure 2: predicted values from model with logit transformation */
graph twoway scatter premeals1 api99, yline(0 1) msym(oh)
/* figure 2: predicted values from model with logit transformation */
graph twoway scatter premeals1 api99, yline(0 1) msym(oh)
 /* figure 3: predicted values from model without transformation */
graph twoway scatter preols api99, yline(0 1) msym(oh)
/* figure 3: predicted values from model without transformation */
graph twoway scatter preols api99, yline(0 1) msym(oh)
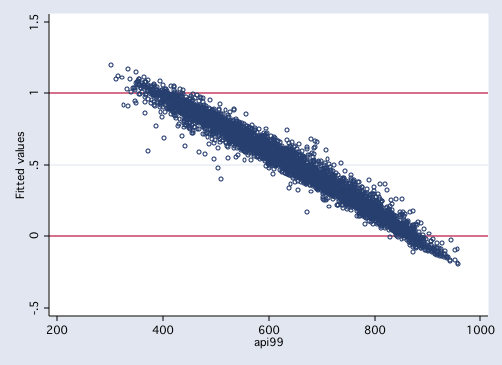
Note that the values from figures 1 and 2 fall within the range of zero to one while those
in figure 3 the values go beyond those bounds.
Let's finish by looking a the correlations of the predicted values with the dependent
variable, meals.| meals premea~1 preols -------------+--------------------------- meals | 1.0000 premeals1 | 0.9152 1.0000 preols | 0.9091 0.9891 1.0000 Note that the correlation between meals and premeals1 is slightly higher than for meals and preols.corr meals premeals1 preols (obs=4257)
Predicting specific values
Now, let's say that you want predicted proportions for some specific combinations of your predictor variables. Specifically, for 500, 600 and 700 for api99, for 1 and 2 for yr_rnd, and for parentrd of 2.5. You would append the following six observations to your dataset with an n of 4421.count
4421
set obs 4427
obs was 4421, now 4427
replace api99 = 500 in 4422
replace api99 = 600 in 4423
replace api99 = 700 in 4424
replace api99 = 500 in 4425
replace api99 = 600 in 4426
replace api99 = 700 in 4427
replace yr_rnd = 1 in 4422/4424
replace yr_rnd = 2 in 4425/4427
replace parented = 2.5 in 4422/4427
list api99 yr_rnd parented in -6/l, separator(3)
+---------------------------+
| api99 yr_rnd parented |
|---------------------------|
4422. | 500 No 2.5 |
4423. | 600 No 2.5 |
4424. | 700 No 2.5 |
|---------------------------|
4425. | 500 Yes 2.5 |
4426. | 600 Yes 2.5 |
4427. | 700 Yes 2.5 |
+---------------------------+
Rerun your model for the 'real' observations (note the in 1/4421), predict for all observations, and display your results.
glm meals yr_rnd parented api99 in 1/4421, link(logit) family(binomial) robust nolog
Generalized linear models No. of obs = 4257
Optimization : ML Residual df = 4253
Scale parameter = 1
Deviance = 395.8141242 (1/df) Deviance = .093067
Pearson = 374.7025759 (1/df) Pearson = .0881031
Variance function: V(u) = u*(1-u/1) [Binomial]
Link function : g(u) = ln(u/(1-u)) [Logit]
AIC = .7220973
Log pseudolikelihood = -1532.984106 BIC = -35143.61
------------------------------------------------------------------------------
| Robust
meals | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
yr_rnd | .0482527 .0321714 1.50 0.134 -.0148021 .1113074
parented | -.7662598 .0390715 -19.61 0.000 -.8428386 -.6896811
api99 | -.0073046 .0002156 -33.89 0.000 -.0077271 -.0068821
_cons | 6.75343 .0896767 75.31 0.000 6.577667 6.929193
------------------------------------------------------------------------------
predict premeals
(option mu assumed; predicted mean meals)
(164 missing values generated)
list api99 yr_rnd parented premeals in -6/l, separator(3)
+--------------------------------------+
| api99 yr_rnd parented premeals |
|--------------------------------------|
4422. | 500 No 2.5 .774471 |
4423. | 600 No 2.5 .6232278 |
4424. | 700 No 2.5 .4434458 |
|--------------------------------------|
4425. | 500 Yes 2.5 .7827873 |
4426. | 600 Yes 2.5 .6344891 |
4427. | 700 Yes 2.5 .4553849 |
+--------------------------------------+