Introduction
Missing data is a common issue, and more often than not, we deal with the matter of missing data in an ad hoc fashion. The purpose of this seminar is to discuss commonly used techniques for handling missing data and common issues that could arise when these techniques are used. In particular, we will focus on the one of the most popular methods, multiple imputation. We are not advocating in favor of any one technique to handle missing data and depending on the type of data and model you will be using, other techniques such as direct maximum likelihood may better serve your needs. We have chosen to explore multiple imputation through an examination of the data, a careful consideration of the assumptions needed to implement this method and a clear understanding of the analytic model to be estimated. We hope this seminar will help you to better understand the scope of the issues you might face when dealing with missing data using this method.
The data set hsb2_mar.dta which is based on hsb2. The Stata code for this seminar is developed using Stata 15. Some of the variables have value labels associated with them.
Goals of statistical analysis with missing data:
- Minimize bias
- Maximize use of available information
- Obtain appropriate estimates of uncertainty
Exploring missing data mechanisms
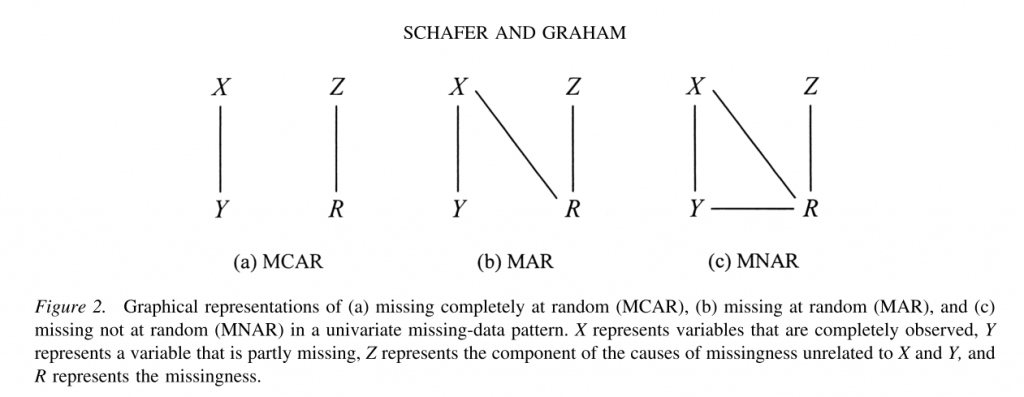
The missing data mechanism describes the process that is believed to have generated the missing values. Missing data mechanisms generally fall into one of three main categories. There are precise technical definitions for these terms in the literature; the following explanation necessarily contains simplifications.
- Missing completely at random (MCAR)
A variable is missing completely at random, if neither the variables in the dataset nor the unobserved value of the variable itself predict whether a value will be missing. Missing completely at random is a fairly strong assumption and may be relatively rare. One relatively common situation in which data are missing completely at random occurs when a subset of cases is randomly selected to undergo additional measurement, this is sometimes referred to as “planned missing.” For example, in some health surveys, some subjects are randomly selected to undergo more extensive physical examination; therefore only a subset of participants will have complete information for these variables. Missing completely at random also allow for missing on one variable to be related to missing on another, e.g. var1 is missing whenever var2 is missing. For example, a husband and wife are both missing information on height.
- Missing at random (MAR)
A variable is said to be missing at random if other variables (but not the variable itself) in the dataset can be used to predict missingness on a given variable. For example, in surveys, men may be more likely to decline to answer some questions than women (i.e., gender predicts missingness on another variable). MAR is a less restrictive assumption than MCAR. Under this assumption the probability of missingness does not depend on the true values after controlling for the observed variables. MAR is also related to ignorability. The missing data mechanism is said be ignorable if it is missing at random and the probability of a missingness does not depend on the missing information itself. The assumption of ignorability is needed for optimal estimation of missing information and is a required assumption for both of the missing data techniques we will discuss.
- Missing not at random (MNAR)
Finally, data are said to be missing not at random if the value of the unobserved variable itself predicts missingness. A classic example of this is income. Individuals with very high incomes are more likely to decline to answer questions about their income than individuals with more moderate incomes.
An understanding of the missing data mechanism(s) present in your data is important because different types of missing data require different treatments. When data are missing completely at random, analyzing only the complete cases will not result in biased parameter estimates (e.g., regression coefficients). However, the sample size for an analysis can be substantially reduced, leading to larger standard errors. In contrast, analyzing only complete cases for data that are either missing at random, or missing not at random can lead to biased parameter estimates. Multiple imputation and other modern methods such as direct maximum likelihood generally assumes that the data are at least MAR, meaning that this procedure can also be used on data that are missing completely at random. Statistical models have also been developed for modeling the MNAR processes; however, these model are beyond the scope of this seminar.
For more information on missing data mechanisms please see:
- Allison, 2002
- Enders, 2010
- Little & Rubin, 2002
- Rubin, 1976
- Schafer & Graham, 2002
Full data:
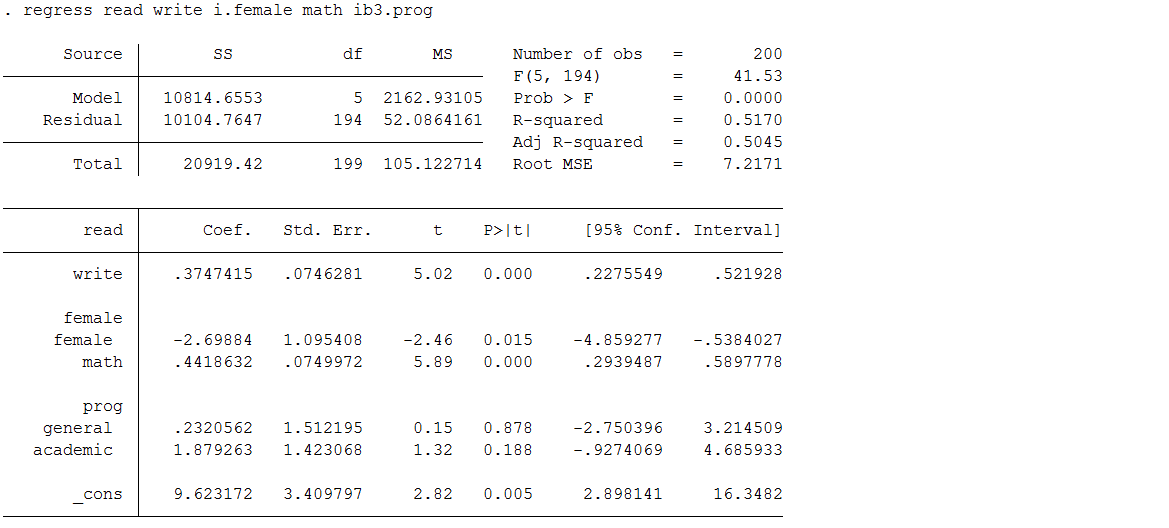
Below is a regression model predicting read using the complete data set (hsb2) used to create hsb_mar, which contains test scores, as well as demographic and school information for 200 high school students. We will use these results for comparison.
use https://stats.idre.ucla.edu/stat/stata/notes/hsb2 regress read write i.female math ib3.prog
Common techniques for dealing with missing data
In this section, we are going to discuss some common techniques for dealing with missing data and briefly discuss their limitations.
- Complete case analysis (listwise deletion)
- Available case analysis (pairwise deletion)
- Mean Imputation
- Single Imputation
- Stochastic Imputation
1. Complete Case Analysis:
This methods involves deleting cases in a particular dataset that are missing data on any variable of interest. It is a common technique because it is easy to implement and works with any type of analysis.
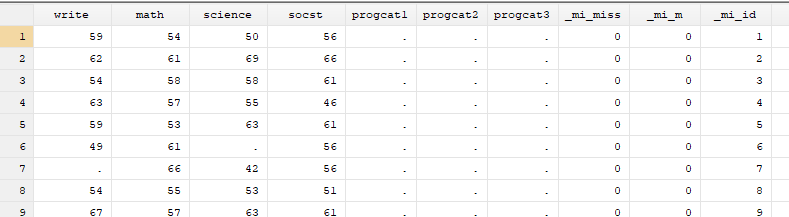
Below we look at some of the descriptive statistics of the data set hsb_mar.
use https://stats.idre.ucla.edu/wp-content/uploads/2017/05/hsb2_mar.dta, clearsum
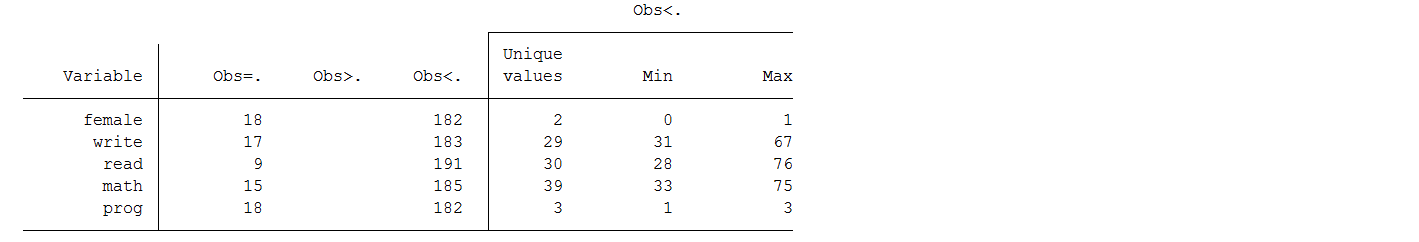
Note that although the dataset contains 200 cases, six of the variables have fewer than 200 observations. The missing information varies between 9 observations or 4.5% (read) and 18 observations or 9% (female and prog) of cases depending on the variable. This doesn’t seem like a lot of missing data, so we might be inclined to try to analyze the observed data as they are, a strategy sometimes referred to as complete case analysis.
Below is a regression model where the dependent variable read is regressed on write, math, female and prog. Notice that the default behavior of the command regress is complete case analysis (also referred to as listwise deletion).
use https://stats.idre.ucla.edu/wp-content/uploads/2017/05/hsb2_mar.dta, clearregress read write i.female math ib3.prog
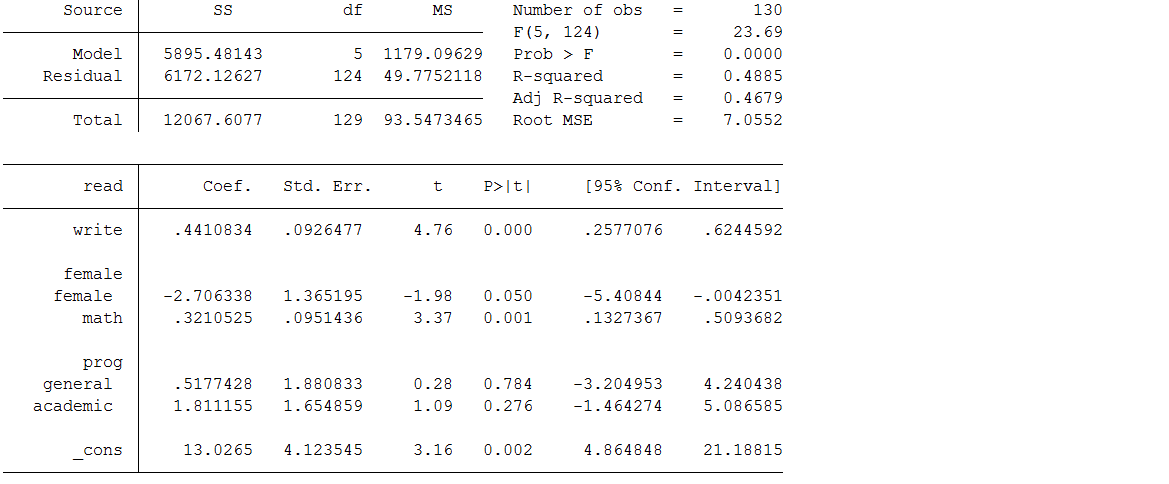
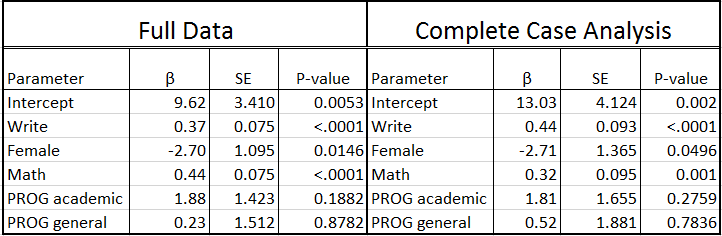
Looking at the output, we see that only 130 cases were used in the analysis; in other words, more than one third of the cases in our dataset (70/200) were excluded from the analysis because of missing data. The reduction in sample size (and statistical power) alone might be considered a problem, but complete case analysis can also lead to biased estimates. Specifically you will see below that the estimates for the intercept, write, math and prog are different from the regression model on the complete data. Also, the standard errors are all larger due to the smaller sample size, resulting in the parameter estimate for female becoming borderline non-significant. Unfortunately, unless the mechanism of missing data is MCAR, this method will introduce bias into the parameter estimates.
2. Available Case Analysis:
This method involves estimating means, variances and covariances based on all available non-missing cases. Meaning that a covariance (or correlation) matrix is computed where each element is based on the full set of cases with non-missing values for each pair of variables. This method became popular because the loss of power due to missing information is not as substantial as with complete case analysis. Depending on the pairwise comparisons examined, the sample size will change based on the amount of missing present in one or both variables. One of the main drawbacks of this method is no consistent sample size and the parameter estimates produced are often much different than the estimates obtained from analysis on the full data or the listwise deletion approach. Unless the mechanism of missing data is MCAR, this method will introduce bias into the parameter estimates. Therefore, this method is not recommended.
3. Unconditional Mean Imputation:
This methods involves replacing the missing values for an individual variable with its overall estimated mean from the available cases. While this is a simple and easily implemented method for dealing with missing values it has some unfortunate consequences. The most important problem with mean imputation, also called mean substitution, is that it will result in an artificial reduction in variability due to the fact you are imputing values at the center of the variable’s distribution. This also has the unintended consequence of changing the magnitude of correlations between the imputed variable and other variables. We can demonstrate this phenomenon in our data.
Below are tables of the means and standard deviations of the four variables in our regression model BEFORE and AFTER a mean imputation as well as their corresponding correlation matrices. This will require us to create dummy variables for our categorical predictor prog.
tab prog, gen(progcat)
Means and correlations between variables before mean imputation
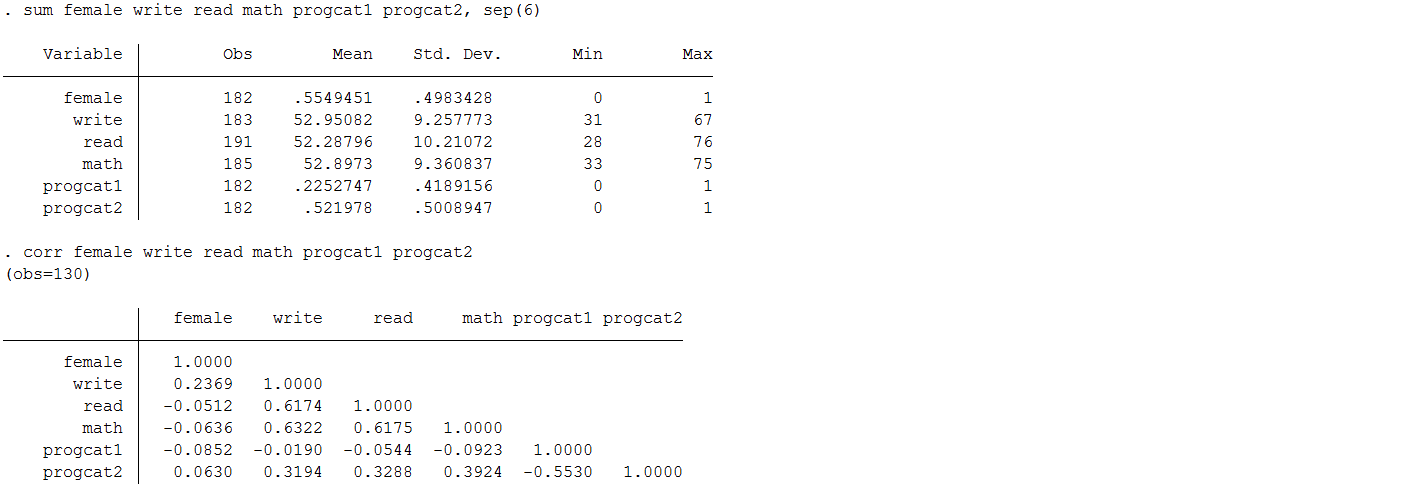
Means and correlations between variables after mean imputation.
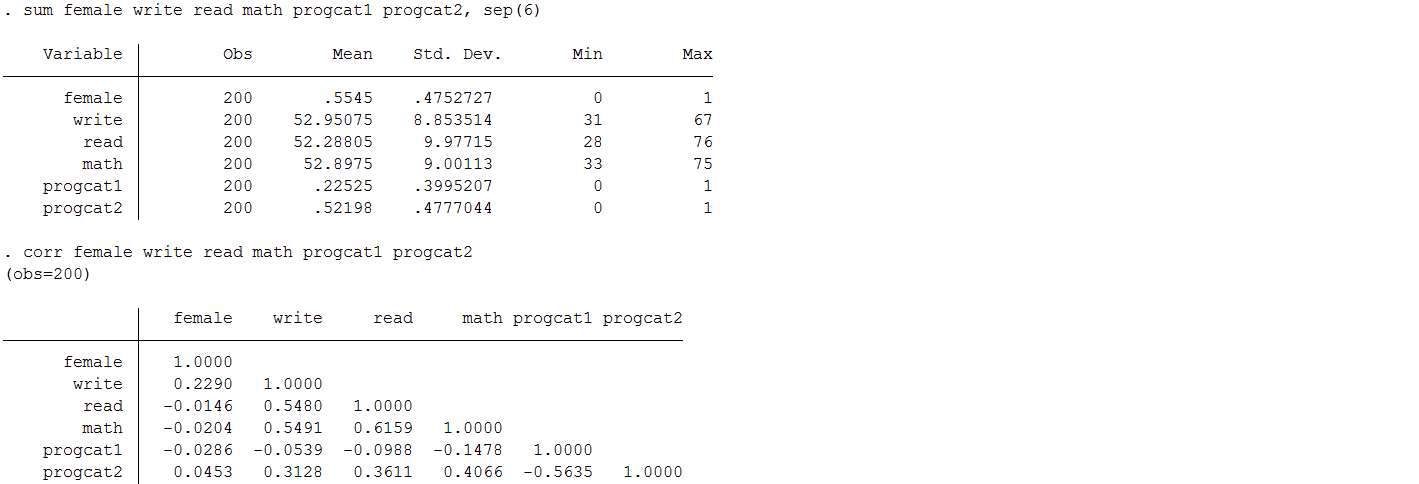
You will notice that there is very little change in the mean (as you would expect); however, the standard deviation is noticeably lower after substituting in mean values for the observations with missing information. This is because you reduce the variability in your variables when you impute everyone at the mean. Moreover, you can see the table of correlation coefficients that the correlation between each of our predictors of interest ( write , math , female , and prog) as well as between predictors and the outcome read have now be attenuated. Therefore, regression models that seek to estimate the associations between these variables will also see their effects weakened.
4. Single or Deterministic Imputation :
A slightly more sophisticated type of imputation is a regression/conditional mean imputation, which replaces missing values with predicted scores from a regression equation. The strength of this approach is that it uses complete information to impute values. The drawback here is that all your predicted values will fall directly on the regression line once again decreasing variability, just not as much as with unconditional mean imputation. Moreover, statistical models cannot distinguish between observed and imputed values and therefore do not incorporate into the model the error or uncertainly associated with that imputed value. Additionally, you will see that this method will also inflate the associations between variables because it imputes values that are perfectly correlated with one another. Unfortunately, even under the assumption of MCAR, regression imputation will upwardly bias correlations and R-squared statistics. Further discussion and an example of deterministic imputation can be found in Craig Enders book “Applied Missing Data Analysis” (2010).
5. Stochastic Imputation :
In recognition of the problems with regression imputation and the reduced variability associated with this approach, researchers developed a technique to incorporate or “add back” lost variability. A residual term, that is randomly drawn from a normal distribution with mean zero and variance equal to the residual variance from the regression model, is added to the predicted scores from the regression imputation thus restoring some of the lost variability. This method is superior to the previous methods as it will produce unbiased coefficient estimates under MAR. However, the standard errors produced during regression estimation while less biased than the single imputation approach, will still be attenuated.
While you might be inclined to use one of these more traditional methods, consider this statement: “Missing data analyses are difficult because there is no inherently correct methodological procedure. In many (if not most) situations, blindly applying maximum likelihood estimation or multiple imputation will likely lead to a more accurate set of estimates than using one of the [previously mentioned] missing data handling techniques” (p.344, Applied Missing Data Analysis, 2010).
Multiple Imputation
Multiple imputation is essentially an iterative form of stochastic imputation. However, instead of filling in a single value, the distribution of the observed data is used to estimate multiple values that reflect the uncertainty around the true value. These values are then used in the analysis of interest, such as in a OLS model, and the results combined. Each imputed value includes a random component whose magnitude reflects the extent to which other variables in the imputation model cannot predict its true values (Johnson and Young, 2011; White et al, 2010). Thus, building into the imputed values a level of uncertainty around the “truthfulness” of the imputed values.
A common misconception of missing data methods is the assumption that imputed values should represent “real” values. The purpose when addressing missing data is to correctly reproduce the variance/covariance matrix we would have observed had our data not had any missing information.
MI has three basic phases:
1. Imputation or Fill-in Phase: The missing data are filled in with estimated values and a complete data set is created. This process of fill-in is repeated m times.
2. Analysis Phase: Each of the m complete data sets is then analyzed using a statistical method of interest (e.g. linear regression).
3. Pooling Phase: The parameter estimates (e.g. coefficients and standard errors) obtained from each analyzed data set are then combined for inference.
The imputation method you choose depends on the pattern of missing information as well as the type of variable(s) with missing information.
Imputation Model, Analytic Model and Compatibility :
When developing your imputation model, it is important to assess if your imputation model is “congenial” or consistent with your analytic model. Consistency means that your imputation model includes (at the very least) the same variables that are in your analytic or estimation model. This includes any transformations to variables that will be needed to assess your hypothesis of interest. This can include log transformations, interaction terms, or recodes of a continuous variable into a categorical form, if that is how it will be used in later analysis. The reason for this relates back to the earlier comments about the purpose of multiple imputation. Since we are trying to reproduce the proper variance/covariance matrix for estimation, all relationships between our analytic variables should be represented and estimated simultaneously. Otherwise, you are imputing values assuming they have a correlation of zero with the variables you did not include in your imputation model. This would result in underestimating the association between parameters of interest in your analysis and a loss of power to detect properties of your data that may be of interest such as non-linearities and statistical interactions. For additional reading on this particular topic see:
1. von Hippel, 2009
2. von Hippel, 2013
3. White et al., 2010
Preparing to conduct MI:
First step: Examine the number and proportion of missing values among your variables of interest. Let’s reload our dataset and use the mdesc command to count the number of missing observations and proportion of missing for each variable.
use https://stats.idre.ucla.edu/wp-content/uploads/2017/05/hsb2_mar.dta, clearmdesc female write read math prog

We can see that the variables with the highest proportion of missing information are prog and female with 9.0%. In general, you want to note the variable(s) with a high proportion of missing information as they will have the greatest impact on the convergence of your specified imputation model.
Second Step: Examine Missing Data Patterns among your variables of interest.
Stata has a suite of multiple imputation (mi) commands to help users not only impute their data but also explore the patterns of missingness present in the data.
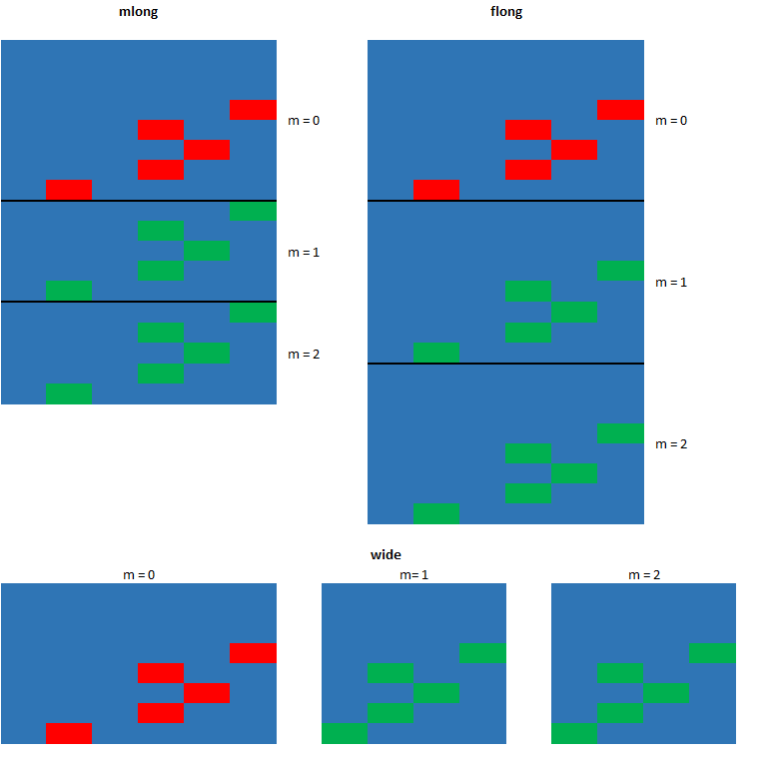
In order to use these commands the dataset in memory must be declared or mi set as “mi” dataset. A dataset that is mi set is given an mi style. This tells Stata how the multiply imputed data is to be stored once the imputation has been completed. For information on these style type help mi styles into the command window. We will use the style mlong.
The chosen style can be changed using mi convert.
mi set mlong
You will notice that executing the previous comand will create three new variables to your dataset. These new variables will be used by Stata to track the imputed datasets and values.
- _mi_miss: marks the observations in the original dataset that have missing values.
- _mi_m: indicates the imputation number. The value is 0 for the original dataset.
- _mi_id: indicator for the observations in the original dataset and is repeated across imputed dataset to mark the imputed observations.
The mi misstable commands helps users tabulate the amount of missing in their variables of interest (summarize) as well as examine patterns of missing (patterns).
mi misstable summarize female write read math prog
Notice that Stata codes missing values ., .a, .b, .c, …, .z as larger than any nonmissing values:
∞ < . < .a < .b < … < .z
mi misstable patterns female write read math prog

This “Missing-value patterns” table is shown above. Each row represents a set of observations in the data set that share the same pattern of missing information. For example, row 1 represents the 65% of observations (n=130) in the data that have complete information on all 5 variables of interest. You can see that there are a total of 12 patterns for the specified variables. You will want to examine this table for any patterns and the appearance of any set of variables that appear to always be missing together. Moreover, depending on the nature of the data, you may also recognize patterns such as monotone missing which can be observed in longitudinal data when an individual drops out at a particular time point and therefore all data after that is subsequently missing. Additionally, you may identify skip patterns that were missed in your original review of the data that should then be dealt with before moving forward with the multiple imputation.
Third Step: If necessary, identify potential auxiliary variables
Auxiliary variables are variables in your data set that are either correlated with a missing variable(s) (the recommendation is r > 0.4) or are believed to be associated with missingness. These are factors that are not of particular interest in your analytic model , but they are added to the imputation model to increase power and/or to help make the assumption of MAR more plausible. These variables have been found to improve the quality of imputed values generate from multiple imputation. Moreover, research has demonstrated their particular importance when imputing a dependent variable and/or when you have variables with a high proportion of missing information (Johnson and Young, 2011; Young and Johnson, 2010; Enders , 2010).
You may a priori know of several variables you believe would make good auxiliary variables based on your knowledge of the data and subject matter. Additionally, a good review of the literature can often help identify them as well. However, if you are not sure what variables in the data would be potential candidates (this is often the case when conducting secondary data analysis), you can uses some simple methods to help identify potential candidates. One way to identify these variables is by examining associations between write, read, female, and math with other variables in the dataset. For example, let’s take a look at the correlation matrix between our 4 variables of interest and two other test score variables science and socst.
tab prog, gen(progcat)
pwcorr female write read math progcat1 progcat2 socst science, obs
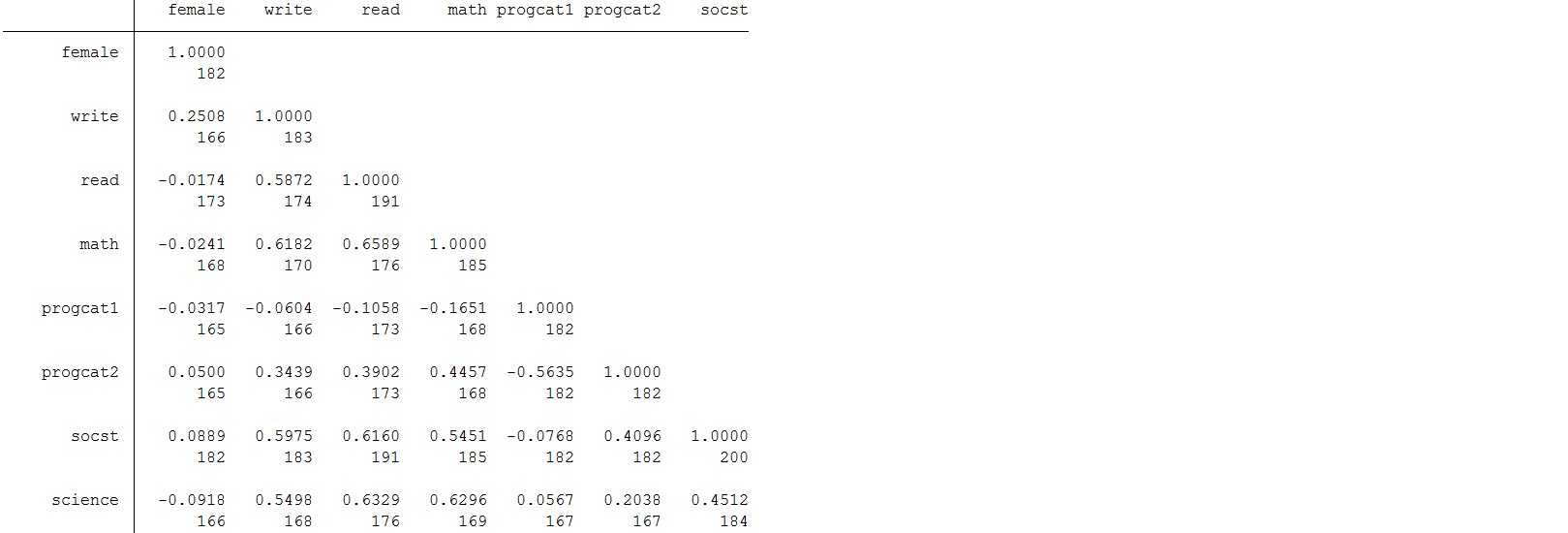
Science and socst both appear to be a good auxiliary because they are well correlated (r >0.4) with all the other test score variables of interest. You will also notice that they are not well correlated with female. A good auxiliary does not have to be correlated with every variable to be useful. You will also notice that science also has missing information of its own. Additionally, a good auxiliary is not required to have complete information to be valuable. They can have missing and still be effective in reducing bias (Enders, 2010).
One area, this is still under active research, is whether it is beneficial to include a variable as an auxiliary if it does not pass the 0.4 correlation threshold with any of the variables to be imputed. Some researchers believe that including these types of items introduces unnecessary error into the imputation model (Allison, 2012), while others do not believe that there is any harm in this practice (Ender, 2010). Thus. we leave it up to you as the researcher to use your best judgment.
Good auxiliary variables can also be correlates or predictors of missingness. Let’s create a set of missing data flags for each variable to be imputed. We will then examine if our potential auxiliary variable socst also appears to predict missingness.
gen female_flag=1 replace female_flag=0 if female==. gen write_flag=1 replace write_flag=0 if write==. gen read_flag=1 replace read_flag=0 if read==. gen math_flag=1 replace math_flag=0 if math==. gen prog_flag=1 replace prog_flag=0 if prog==.
Below are a set of t-tests to test if the mean socst or science scores differ significantly between those with missing information and those without.
foreach var of varlist female_flag - prog_flag{
display "`var'"
ttest socst, by(`var')
}
foreach var of varlist female_flag - prog_flag{
display "`var'"
ttest science, by(`var')
}
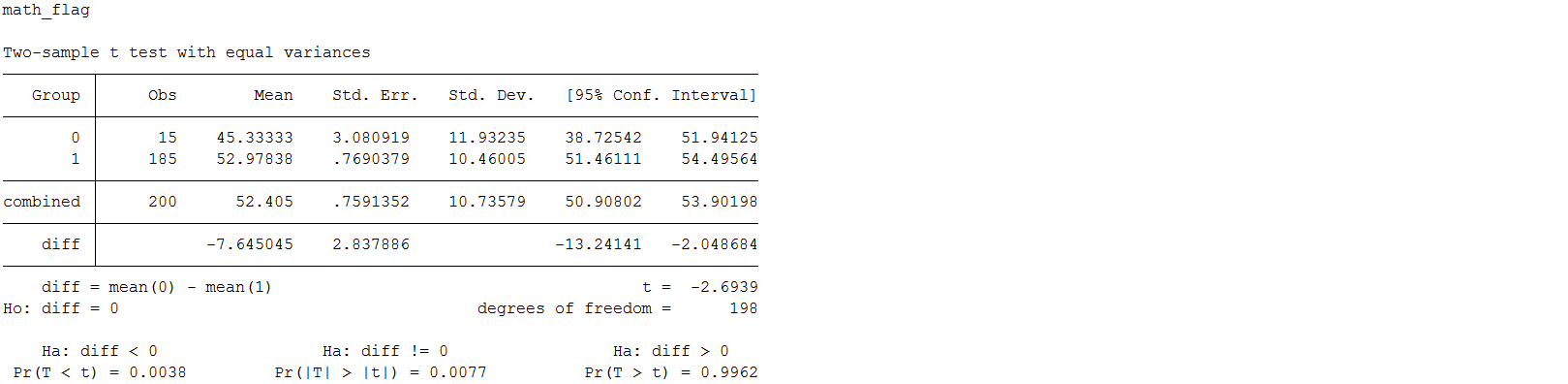
The only significant difference was found when examining missingness on math with socst. Above you can see that the mean socst score is significantly lower among the respondents who are missing on math. This suggests that socst is a potential correlate of missingness (Enders, 2010) and may help us satisfy the MAR assumption for multiple imputation by including it in our imputation model.
Example 1: MI using multivariate normal distribution (MVN):
Upon choosing to impute one or many variables, one of the first decisions you will make is the type of distribution under which you want to impute your variable(s). One available method uses Markov Chain Monte Carlo (MCMC) procedures which assume that all the variables in the imputation model have a joint multivariate normal distribution. This is probably the most common parametric approach for multiple imputation. The specific algorithm used is called the data augmentation (DA) algorithm, which belongs to the family of MCMC procedures. The algorithm fills in missing data by drawing from a conditional distribution, in this case a multivariate normal, of the missing data given the observed data. In most cases, simulation studies have shown that assuming a MVN distribution leads to reliable estimates even when the normality assumption is violated given a sufficient sample size (Demirtas et al., 2008; KJ Lee, 2010). However, biased estimates have been observed when the sample size is relatively small and the fraction of missing information is high.
Note: Since we are using a multivariate normal distribution for imputation, decimal and negative values are possible. These values are not a problem for estimation; however, we will need to create dummy variables for the nominal categorical variables so the parameter estimates for each level can be interpreted.
1. Imputation Phase:
After the data is mi set, Stata requires 3 additional commands. The first is mi register imputed. This command identifies which variables in the imputation model have missing information.
mi register imputed female write read math progcat1 progcat2 science
The second command is mi impute mvn where the user specifies the imputation model to be used and the number of imputed datasets to be created.
mi impute mvn female write read math progcat1 progcat2 science = socst, add(10) rseed (53421)
On the mi impute mvn command line we can use the add option to specify the number of imputations to be performed. In this example we chose 10 imputations. Variables on the left side of the equal sign have missing information, while the right side is reserved for variables with no missing information and are therefore solely considered “predictors” of missing values. As you can see, even through science is an auxiliary variable, science must be included as a variable to be imputed.
After the mvn all the variables for the imputation model are specified including all the variables in the analytic model as well as any auxiliary variables. The option rseed is not required, but since MI is designed to be a random process, setting a seed will allow you to obtain the same imputed dataset each time. The imputed datasets will be stored appended or “stacked” together in a dataset. The indicator variable called _mi_m is automatically populated to number each new imputed dataset (1 -10).

The output after mi impute mvn, let’s the user know what options have been invoked for the command. The top of the output shows what type of imputation was used (MVN), as well as the number of imputed data sets created (m=10). Because the estimation of the imputed values involves a Bayesian process, characteristics of the MCMC are also reported, including the type of prior used, the total number of iterations, the number of burn-in iterations (number of iterations before the first set of imputed values is drawn) and the number of iterations between draws. The bottom portion of the output includes a table that reports the number of missing values that were imputed for each variable that was registered to be imputed.
2. Analysis/Pooling Phase:
The third step is mi estimate which runs the analytic model of interest (here it is a linear regression using regress) within each of the imputed datasets. It also combines all the estimates (coefficients and standard errors) across all the imputed datasets.
The mi estimate command is used as a prefix to the standard regress command. This executes the specified estimation model within each of the 10 imputed datasets to obtain 10 sets of coefficients and standard errors. Stata then combines these estimates to obtain one set of inferential statistics.
mi estimate: regress read write female math progcat1 progcat2

This step combines the parameter estimates into a single set of statistics that appropriately reflect the uncertainty associated with the imputed values. The regression coefficients are simply just an arithmetic mean of the individual coefficients estimated for each of the 10 regression models. Averaging the parameter estimates dampens the variation thus increasing efficiency and decreasing sampling variation. Estimation of the standard error for each variable is little more complicated and will be discussed in the next section. If you compare these estimates to those from the complete data you will observe that they are, in general, quite comparable. The variables write, female and math, are significant in both sets of data. You will also observe a small inflation in the standard errors, which is to be expected since the multiple imputation process is designed to build additional uncertainty into our estimates.
| Full Data | Complete Case | MVN Imputation | |||||||||
| Parameter | β | SE | P-value | Parameter | β | SE | P-value | Parameter | β | SE | P-value |
| Intercept | 9.62 | 3.410 | 0.0053 | Intercept | 13.03 | 4.124 | 0.002 | Intercept | 10.35 | 3.687 | 0.006 |
| Write | 0.37 | 0.075 | <.0001 | Write | 0.44 | 0.093 | <.0001 | Write | 0.39 | 0.082 | <.0001 |
| Female | -2.70 | 1.095 | 0.0146 | Female | -2.71 | 1.365 | 0.0496 | Female | -2.74 | 1.144 | 0.017 |
| Math | 0.44 | 0.075 | <.0001 | Math | 0.32 | 0.095 | 0.001 | Math | 0.40 | 0.087 | <.0001 |
| PROG academic | 1.88 | 1.423 | 0.1882 | PROG academic | 1.81 | 1.655 | 0.2759 | PROG academic | 2.81 | 1.602 | 0.083 |
| PROG general | 0.23 | 1.512 | 0.8782 | PROG general | 0.52 | 1.881 | 0.7836 | PROG general | 0.52 | 1.685 | 0.76 |
2. Imputation Diagnostics:
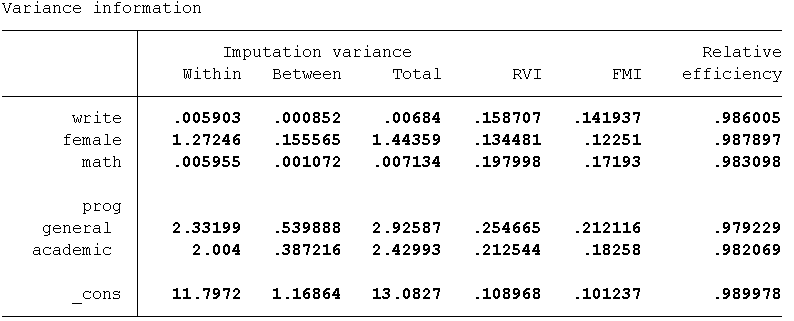
In the output from mi estimate you will see several metrics in the upper right hand corner that you may find unfamilar These parameters are estimated as part of the imputation and allow the user to assess how well the imputation performed. By default, Stata provides summaries and averages of these values but the individual estimates can be obtained using the vartable and dftable options.
Let’s take a look at the information for RVI (Relative Increase in Variance), FMI (Fraction of Missing Information), DF (Degrees of Freedom) , RE (Relative Efficiency), as well as the between imputation and the within imputation variance estimates to examine how the standard errors (SEs) are calculated.
mi estimate, vartable dftable
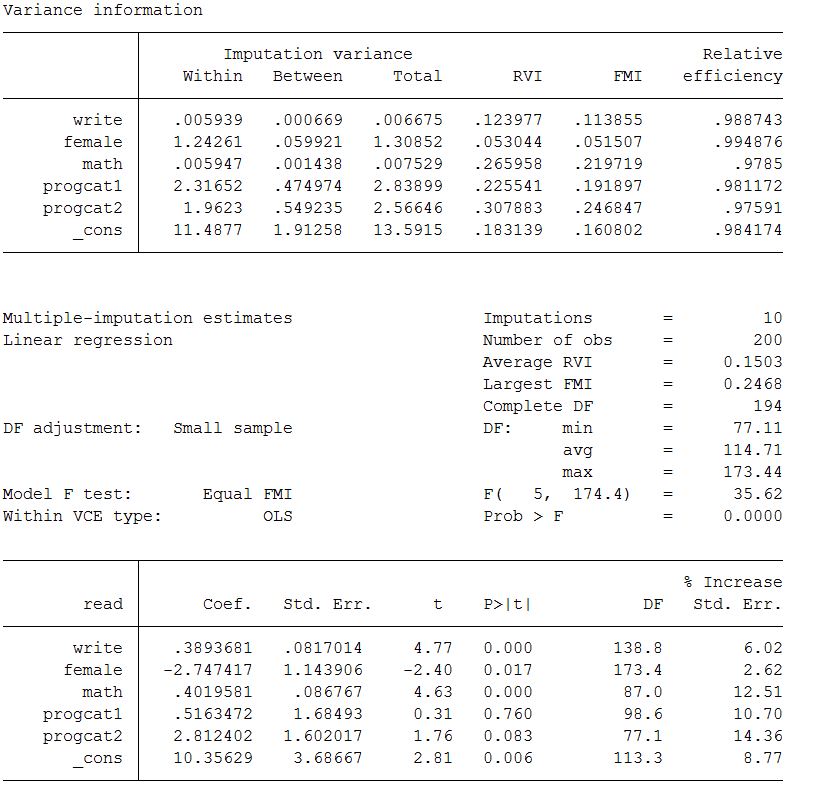
Below we discuss each piece:
- Variance Between (VB):
- This is a measure of the variability in the parameter estimates
(coefficients) obtained from the 10 imputed datasets
- For example, if you took all 10 of the parameter estimates for write and calculated the variance this would equal VB = 0.00067.
- This variability estimates the additional variation (uncertainty) that results from missing data.
- This is a measure of the variability in the parameter estimates
(coefficients) obtained from the 10 imputed datasets
- Variance Within (VW):
- This is simply the arithmetic mean of the sampling
variances (SE) from each of the 10 imputed datasets.
- For example, if you squared the standard errors for write for all 10 imputations and then divided by 10, this would equal Vw = 0.0059
- This estimates the sampling variability that we would have expected had there been no missing data.
- Variance Total (VT):
-
- The primary usefulness of MI comes from how the total variance is estimated.
- The total variance is sum of multiple sources of variance.
- While regression coefficients are just averaged across imputations, Rubin’s formula (Rubin, 1987) partitions variance into “within imputation” capturing the expected uncertainty and “between imputation” capturing the estimation variability due to missing information (Graham, 2007; White et al., 2010).
- The total variance is the sum of 3 sources
of variance. The within, the between and an
additional source of sampling variance.
- For example, the total variance for the variable write would be calculated like this: VB + Vw + VB/m = 0.00067 + 0.0059 + 0.00067/10 = 0.00667
- The additional sampling variance is literally the
variance between divided by m.
This value represents the sampling error associated with the overall or
average coefficient estimates. It is used as a correction factor for
using a specific number of imputations.
- This value becomes smaller, the more imputations are conducted. The idea being that the larger the number of imputations, the more precise the parameter estimates will be.
- Bottom line: The main difference between multiple imputation and other single imputation methods, is in the estimation of the variances. The SEs for each parameter estimate are the square root of their VT s
-
- Relative Increases in Variance (RIV/RVI):
- Proportional increase in total sampling variance that is due to missing information ([VB + VB/m]/VW).
- For example, the RVI for write is 0.1239 this means that the estimated sampling variance for write is 12.4% larger than its sampling variance would have been had the data on write been complete.
- Bottom line: Variables with large amounts of missing and/or that are weakly correlated with other variables in the imputation model will tend to have high RVI’s.
- Fraction of Missing Information (FMI):
- Is directly related to RVI.
- Proportion of the total sampling variance that is due to missing data ([VB+ VB/m ]/VT) .
- It’s estimated based on the percentage missing for a particular variable and how correlated this variable is with other variables in the imputation model.
- The interpretation is similar to an R-squared. So an FMI of 0.1138 for write means that 11.4% of the total sampling variance is attributable to missing data.
- The accuracy of the estimate of FMI increases as the number imputation increases because variance estimates stabalize with larger numbers imputations. This especially important in the presence of a variable(s) with a high proportion of missing information.
- If convergence of your imputation model is slow, examine the FMI estimates for each variables in your imputation model. A high FMI can indicate a problematic variable.
- Bottom line: If FMI is high for any particular variable(s) then consider increasing the number of imputations. A good rule of thumb is to have the number imputations (at least) equal the highest FMI percentage.
- Relative Efficiency:
- The relative (variance) efficiency (RE) of an imputation (how well the true population parameters are estimated) is related to both the amount of missing information as well as the number (m) of imputations performed.
- RE is an estimate of the effficiency relative to performing an infinite number of imputations.
- When the amount of missing information is very low then efficiency may be achieved by only performing a few imputations (the minimum number given in most of the literature is 5). However when there is high amount of missing information, more imputations are typically necessary to achieve adequate efficiency for parameter estimates. You can obtain relatively good efficiency even with a small number of m. However, this does not mean that the standard errors will be well estimated well.
- More imputations are often necessary for proper standard error estimation as the variability between imputed datasets incorporate the necessary amount of uncertainty around the imputed values.
- The direct relationship between RE, m and the FMI is: 1/(1+FMI/m). This formula represent the RE of using m imputation versus the infinte number of imputations. To get an idea of what this looks like practically, take a look at an example from SAS documentation.
- Bottom line: It may appear that you can get good RE with a few imputations; however, it often takes more imputations to get good estimates of the variances than good estimates of parameters like means or regression coefficients.
- Degrees of Freedom (DF):
- Unlike analysis with non-imputed data, sample size does not directly influence the estimate of DF.
- DF actually continues to increase as the number of imputations increase.
- The standard formula used to calculate DF can result in fractional estimates and inflated degrees of freedom.
- A small-sample correction to the DF (Barnard and Rubin, 1999) is implemented (by default) in order to address the inflated DF the can sometimes occur when the number of m is large.
- Bottom line: The standard uncorrected formula assumes that the estimator has a normal distribution, i.e. a t-distribution with infinite degrees of freedom. In large samples this is not usually an issue but can be with smaller sample sizes. In that case, the default corrected formula should be used (Lipsitz et al., 2002).
After performing an imputation it is also useful to look at means, frequencies and box plots comparing observed and imputed values to assess if the range appears reasonable. You may also want to examine plots of residuals and outliers for each imputed dataset individually. If anomalies are evident in only a small number of imputations then this indicates a problem with the imputation model (White et al, 2010).
You should also assess convergence of your imputation model. This should be done for different imputed variables, but specifically for those variables with a high proportion of missing (e.g. high FMI). Convergence of the imputation model means that DA algorithm has reached an appropriate stationary posterior distribution. Convergence for each imputed variable can be assessed using trace plots. Trace plots are plots of estimated parameters against iteration numbers. These plots can be requested using the saveptrace and mcmconly option.
This mcmconly option will simply run the MCMC algorithm for the same number of iterations it takes to obtain 10 imputations without actually producing 10 imputed datasets. Is it typically used in combination with saveptrace or savewlf to examine the convergence of the MCMC prior to imputation. No imputation is performed with mcmconly is specified, so the options add or replace are not required with mi impute mvn.
In practice, convergence is often examined visually from the trace and autocorrelation plots of the estimated parameters. Trace plots are plots of estimated parameters against iteration numbers. Long-term trends in trace plots and high serial dependence in autocorrelation plots are indicative of a slow convergence to stationarity. A stationary process has a mean and variance that do not change over time (StataCorp,2017 – Stata 15 “MI Impute Chained”). You can take a look at examples of good and bad trace plots in the SAS users guide section on “Assessing Markov Chain Convergence“.
mi impute mvn write read female math science progcat1 progcat2 = socst, mcmconly burnin(1000) rseed(53421) saveptrace(trace, replace)
Note that the trace file that is saved is not a true Stata dataset, but it can be loaded as if they were using the mi ptrace use command and its contents can be described without actually opening the file using the command mi ptrace describe. The trace file contains information on imputation number, iteration number, regression coefficients, variances and covariances.
mi ptrace describe trace mi ptrace use trace, clear

If you have a lot of parameters in your model it may not be feasible to examine the convergence of each individual parameter. In that case you can use savewlf. WLF stands for worst linear function. This will output to you the parameter(s) with the highest FMI value.
Let’s take a look at the data for female (y3), which was one of the variables that contain the fewest number of complete observations. We will generate graphs for each series.
We will start by declaring the data as time series, so iteration number will be on the x-axis. We will then graph the regression coefficients and variance for female.
tsset itertsline b_y3x1, name(gr1,replace) ytitle("Coefficient on Socst" "used to predict Female") xtitle("iter") tsline v_y3y3, name(gr2,replace) ytitle(Female Variance) xtitle("iter") graph combine gr1 gr2 , xcommon cols(1) b1title(Iteration)
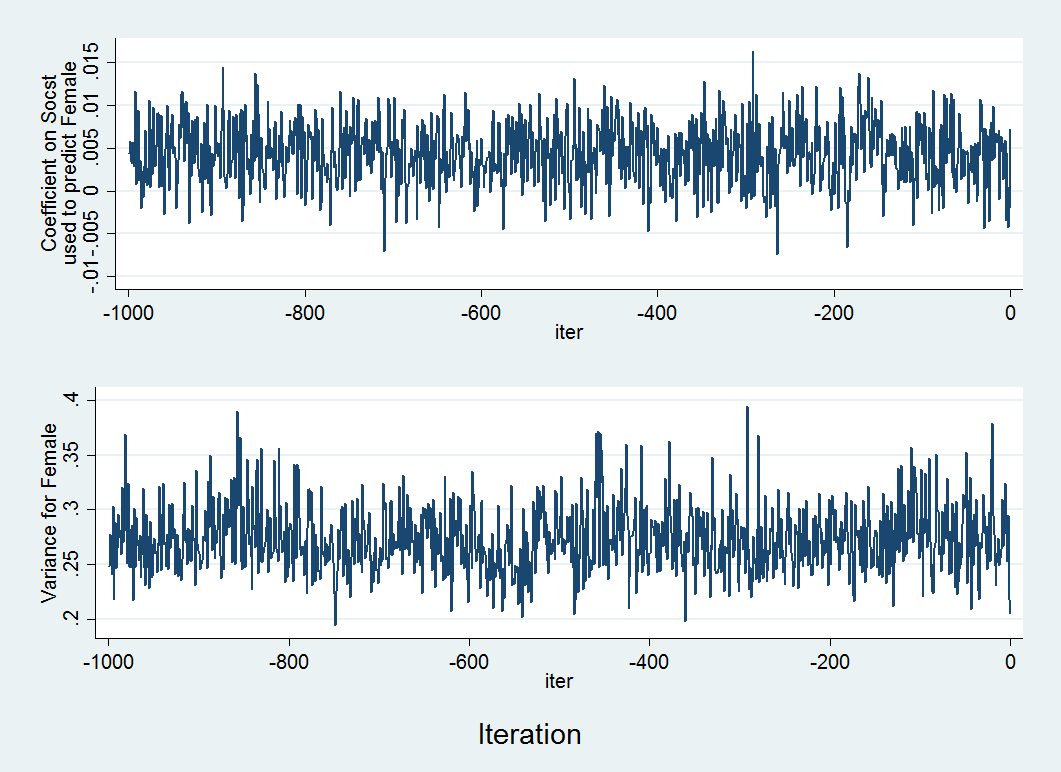
Above is an example of two trace plots. There are two main things you want to note in a trace plot. First, assess whether the algorithm appeared to reach a stable posterior distribution by examining the plot to see if the predicted values remains relatively constant and that there appears to be an absence of any sort of trend (indicating a sufficient amount of randomness in the coefficients, covariances and/or variances between iterations). In our case, this looks to be true. Second, you want to examine the plot to see how long it takes to reach this stationary phase. In the above example it looks to happen almost immediately, as no observable pattern emerges, indicating good convergence. By default the burn-in period (number of iterations before the first set of imputed values is drawn) is 100. This can be increased if it appears that proper convergence is not achieved using the burnin option.
Another plot that is very useful for assessing convergence is the auto correlation plot. Autocorrelation measures the correlation between predicted values at each iteration. As the imputation process os designed to be random, we should not observe correlated imputed values across imputations. We can check to see that enough iterations were left between successive draws (i.e., datasets) that autocorrelation does not exist. Let’s say you noticed a trend in the variances in the previous trace plot. You may want to assess the magnitude of the observed dependency of values across iterations. The auto correlation plot will show you that. To produce these plots in Stata, you will use the ac or autocorrelation command on the same “trace” datafile.
ac b_y3x1, ytitle("Coefficient on Socst" "used to predict Female") xtitle("") ciopts(astyle(none)) ///note("") name(ac1, replace) lags(100)ac v_y3y3, ytitle(Female Variance) xtitle("") ciopts(astyle(none)) note("") name(ac2, replace) lags(100)graph combine ac1 ac2, xcommon cols(1) title(Autocorrelations) b1title(Lag)
In the graph below, the x-axis shows the lag, that is the distance between a given iteration and the iteration it is being correlated with, on the y-axis is the value of the correlations. In the plot you can see that the correlation is high when the mcmc algorithm starts but quickly goes to near zero after a few iterations indicating almost no correlation between iterations and therefore no correlation between values in adjacent imputed datasets. By default Stata, draws an imputed dataset every 100 iterations, if correlation appears high for more than that, you will need to increase the number of iterations between imputed datasets using the burnbetween option. Take a look at the Stata 15 mi impute mvn documentation for more information about this and other options.
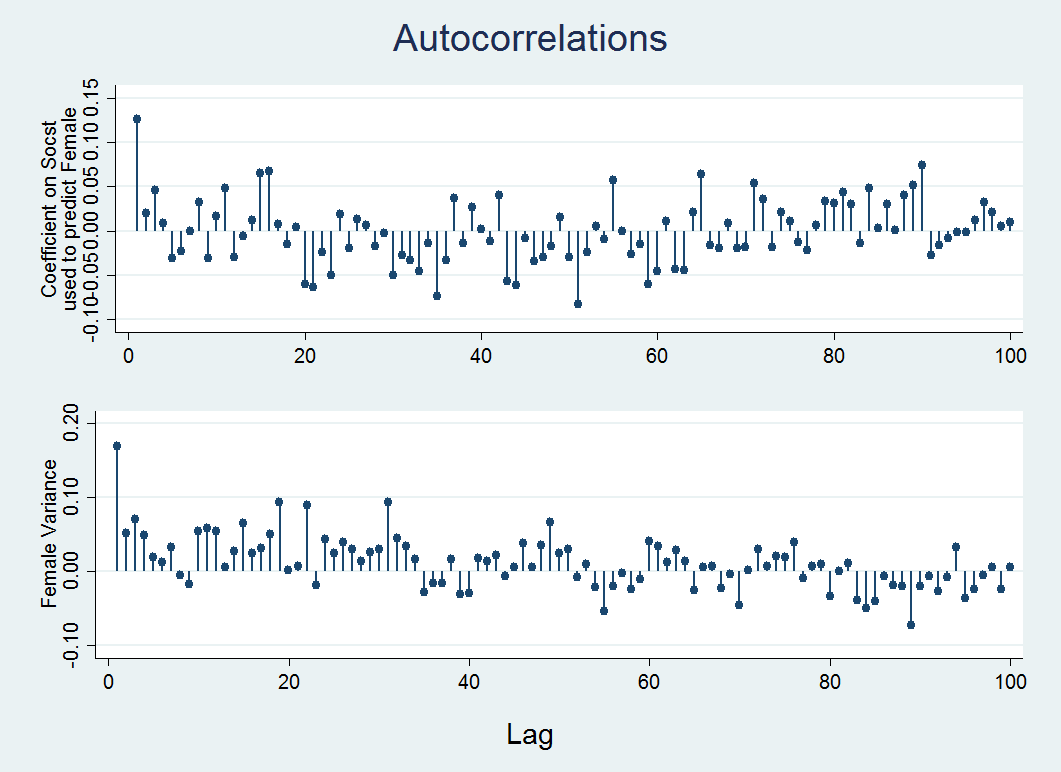
Note: The amount of time it takes to get to zero (or near zero) correlation is an indication of convergence time (Enders, 2010).
For more information on these and other diagnostic tools, please see Ender, 2010 and Rubin, 1987.
Example 2: MI using chained equations/MICE (also known as the fully conditional specification or sequential generalized regression)
A second method available in Stata is multiple imputation by chained equations (MICE) which does not assume a joint MVN distribution but instead uses a separate conditional distribution for each imputed variable. This specification may be necessary if you are are imputing a variable that must only take on specific values such as a binary outcome for a logistic model or a count variable for a Poisson model. In simulation studies (Lee & Carlin, 2010; Van Buuren, 2007), MICE has been show to produce estimates that are comparable to MVN method. Later we will discuss some diagnostic tools that can be used to assess if convergence was reached when using MICE.
The MICE distributions available is Stata are binary, ordered and multinomial logistic regression for categorical variables, linear regression and predictive mean matching (PMM)* for continuous variables, and Poisson and negative binomial regression for count variables. Stata also provides access to some more specialized methods including truncated and interval regression. If you do not specify a distribution, by default, linear regression is used.
For more information on these methods and the options associated with them, see Stata help file on the mi impute chained.
*Note: The default Stata behavior for PMM uses too few nearest neighbor matches and will reuslt sin underestimated stanrds erros, this option should be changed when using the procedure.
Let’s reload the data and mi set:
use https://stats.idre.ucla.edu/wp-content/uploads/2017/05/hsb2_mar.dta, clear mi set mlong
1. Imputation Phase:
The basic set-up for conducting an imputation is shown below. The syntax look very similar to the previous model using MVN with a few differences. First, we are now specifying chained instead of mvn. Second, instead of just listing the variable(s) to be imputed, we will now specify a particular distribution to impute under. The chosen imputation method is listed with parentheses directly preceding the variable(s) to which this distribution applies. In this case, we will use logistic for the binary variable female, multinomial logistic for our unordered categorical variable prog, and linear regression for all of our continuous score variables. You will notice that we no longer need dummy variables for prog since we are imputing it as a categorical variable.
mi register imputed female write read math prog science mi impute chained (logit) female (mlogit) prog (regress) write read math science = socst, ///add(10) rseed (53421) savetrace(trace1,replace)
By default, the variables will be imputed in order from the most observed to the least observed. If you would like to override that default, specify the option orderasis.
As was the case with MVN, Stata will automatically create the variables _mi_m,_mi_id, _mi_miss.
2. Analysis Phase
Once the 10 multiply imputed datasets have been created, we can run our linear regression using the regress command. Since we imputed female and prog under a distribution appropriate for categorical outcomes, the imputed values will now be true integer values and can be treated as indicator variables in a regression model.
mi estimate: regress read write i.female math i.prog
As before, the mi estimate command is used as a prefix to the standard regress command. This executes the specified estimation model on each of the 10 imputed datasets to obtain 10 sets of coefficients and standard errors. Stata then combines these estimates to obtain one set of inferential statistics.
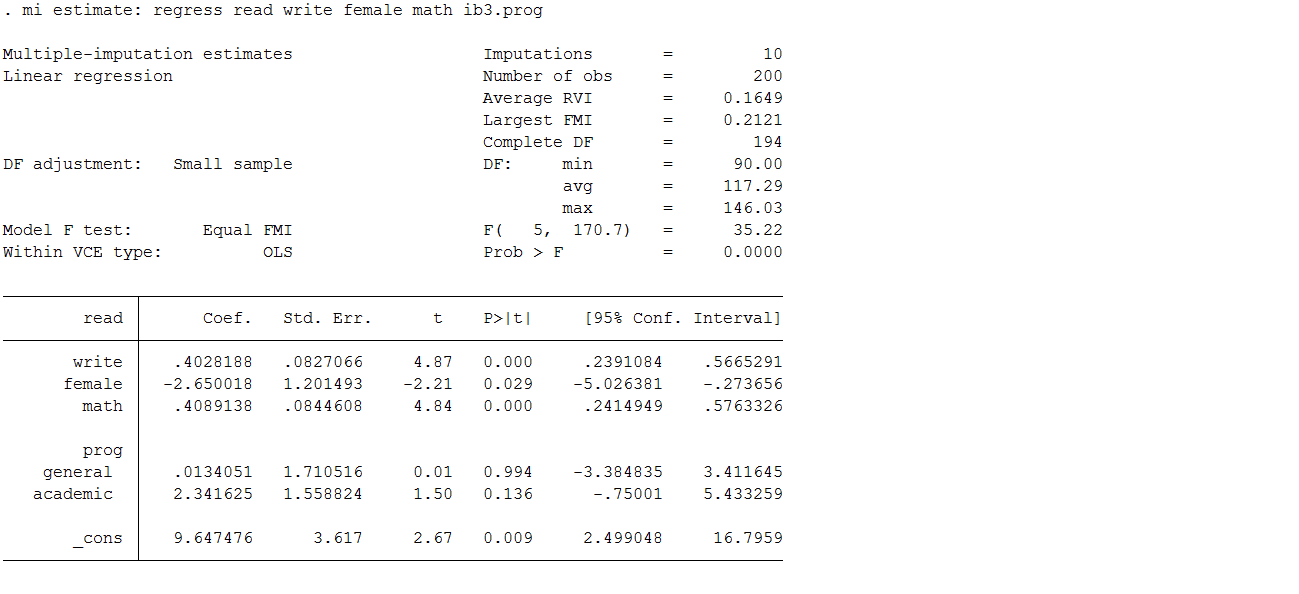
If you compare these estimates to those from the complete data you will observe that they are, in general, quite comparable. The variables write, female and math, are significant in both sets of data. You will also observe a small inflation in the standard errors, which is to be expected since the multiple imputation process is designed to build additional uncertainty into our estimates. As with the MVN model, the SE are larger due to the incorporation of uncertainty around the parameter estimates, but these SE are still smaller than we observed in the complete cases analysis. The parameter estimates all look good except for those for prog.
| Full Data | Complete Case | MICE | |||||||||
| Parameter | β | SE | P-value | Parameter | β | SE | P-value | Parameter | β | SE | P-value |
| Intercept | 9.62 | 3.410 | 0.0053 | Intercept | 13.03 | 4.124 | 0.002 | Intercept | 9.65 | 3.620 | 0.009 |
| Write | 0.37 | 0.075 | <.0001 | Write | 0.44 | 0.093 | <.0001 | Write | 0.40 | 0.083 | <.0001 |
| Female | -2.70 | 1.095 | 0.0146 | Female | -2.71 | 1.365 | 0.0496 | Female | -2.65 | 1.201 | 0.029 |
| Math | 0.44 | 0.075 | <.0001 | Math | 0.32 | 0.095 | 0.001 | Math | 0.41 | 0.084 | <.0001 |
| PROG academic | 1.88 | 1.423 | 0.1882 | PROG academic | 1.81 | 1.655 | 0.2759 | PROG academic | 2.34 | 1.559 | 0.136 |
| PROG general | 0.23 | 1.512 | 0.8782 | PROG general | 0.52 | 1.881 | 0.7836 | PROG general | 0.01 | 1.711 | 0.994 |
Take a look at some of our imputation diagnostic measures and plots to assess if anything needs to be changed about our imputation model. Let’s again examine the RVI, FMI, DF, RE as well as the between imputation and the within imputation variance estimates. As can be seen in the table below, the highest estimated RVI (25%) and FMI (21%) are associated with prog. This indicates that the imputation could potentially be improved by increasing the number of imputations to 20 or 25 as well as including an auxiliary variable(s) associated with prog.
As with the MVN method, we can save a file of the predicted values from each iteration and graph them using a trace plot. The option savetrace specifies Stata to save the means and standard deviations of imputed values from each iteration to a Stata dataset named “trace1”.
Note: When using MVN the option is saveptrace.
use trace1,clear describe

Some data management is necessary in order to create the trace plot. The file produced by Stata is “long” with a row for each chain at each iteration. We want the date “wide” so that the value of mean and standard deviation for each variable are separate by chain. Since there are multiple chains (m=10), iteration number is repeated which is not allowed for time series data. Thus, we need to reshape the data beifre we can use tsset. Then we can graph the predict mean and/or standard deviation for each imputed variable.
reshape wide *mean *sd, i(iter) j(m) tsset itertsline read_mean1, name(mice1,replace)legend(off) ytitle("Mean of Read")
The trace plot below graphs the predicted means value produced during the first imputation chain. As before, the expectations is that the values would vary randomly to incorporate variation into the predicted values for read.
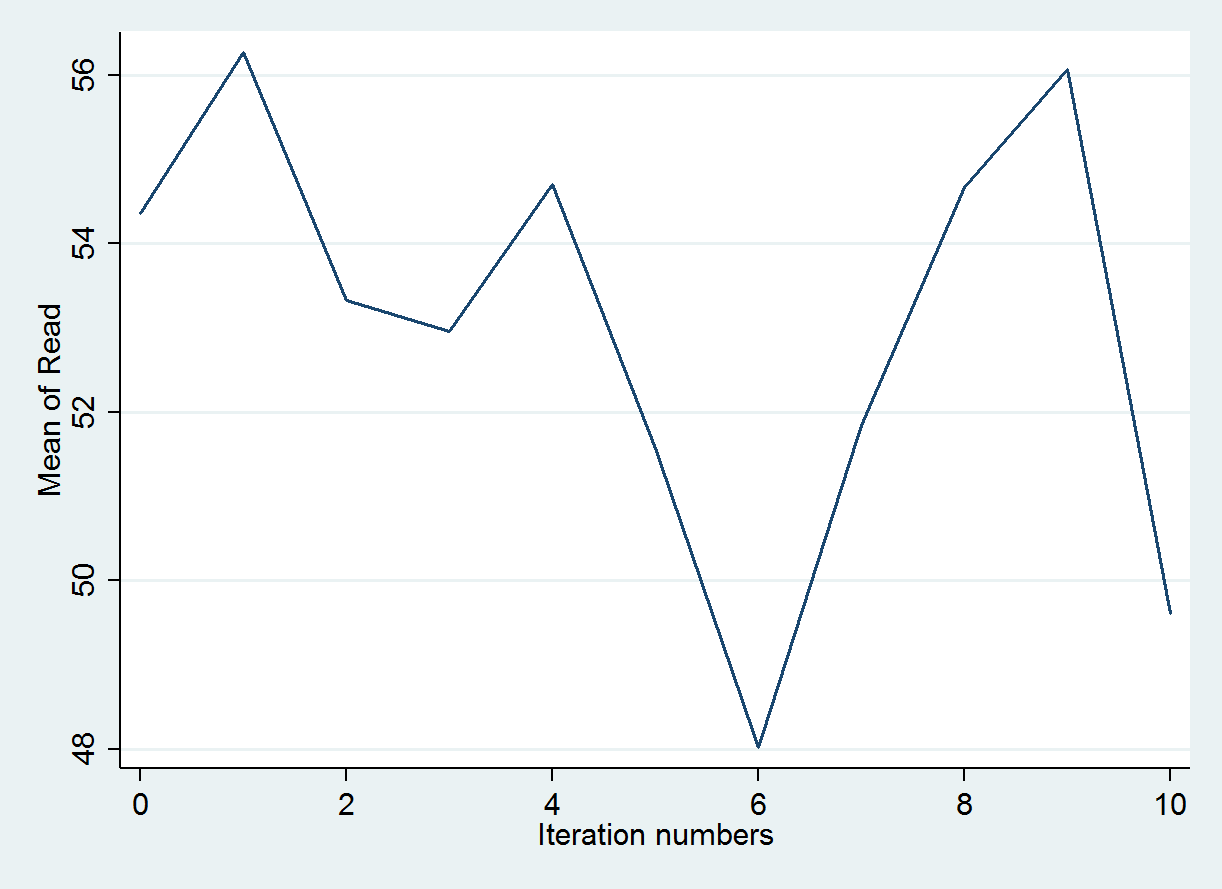
All 10 imputation chains can also be graphed simultaneously to make sure that nothing unexpected occurred in a single chain. Every chain is obtained using a different set of initial values and this should be unique. Each colored line represents a different imputation. So all 10 imputation chains are overlaid on top of one another.
tsline read_mean*, name(mice1,replace)legend(off) ytitle("Mean of Read")
tsline read_sd*, name(mice2, replace) legend(off) ytitle("SD of Read")
graph combine mice1 mice2, xcommon cols(1) title(Trace plots of summaries of imputed values)
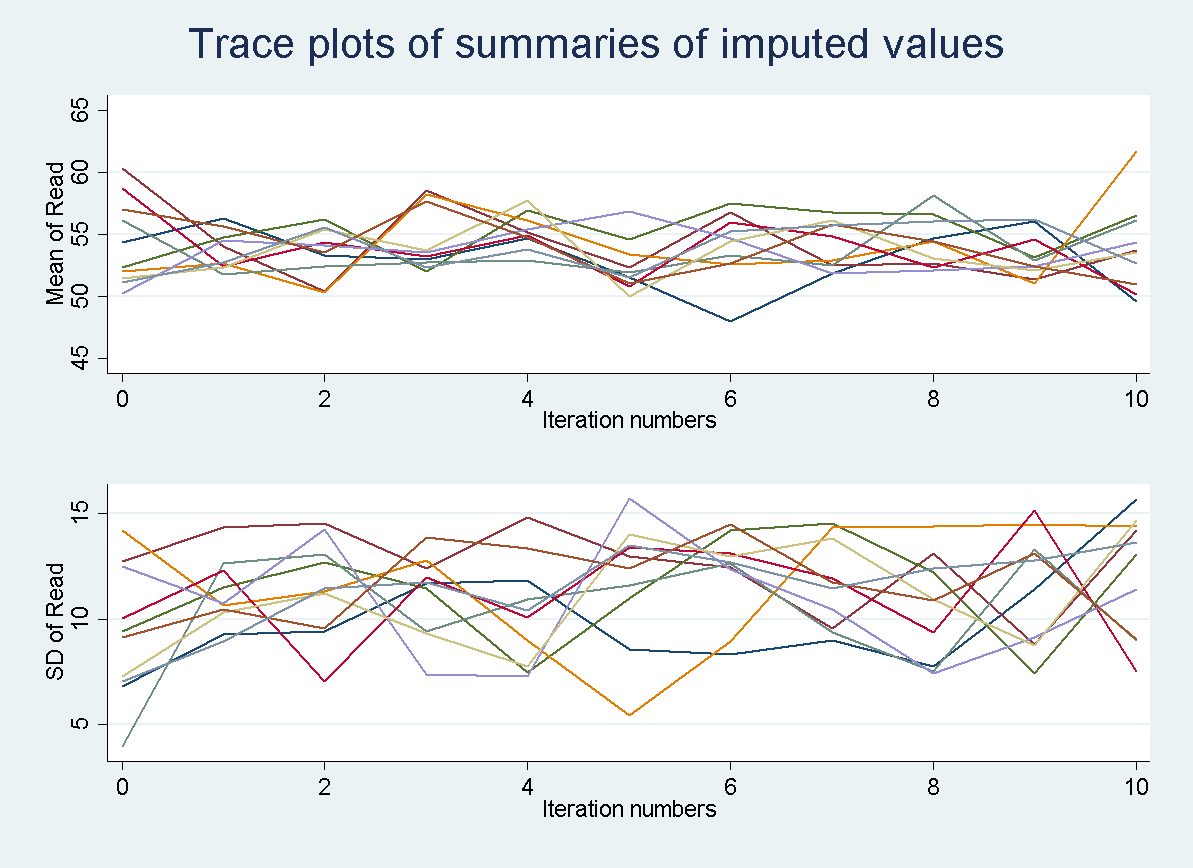
Autocorrelation plots are only available when asuming a joint MVN. In the case of MICE it would have little usefulness due to the iterative process used to create the imputations. In each iteration, the imputation model is estimated using both the observed data and imputed data from the previous iteration. Thus, you will always get a certain amount of autocorrelation. For more information on assessing convergence when using MICE check out Stata’s documentation on mi impute chained.
MICE has several properties that make it an attractive alternative to the DA algorithm. First, the MICE allows each variable to be imputed using its own conditional distribution instead of one common multivariate distribution. This especially useful when negative or non-integer values can not be used in subsequent analyses such as imputing a binary outcome variable. Second, different imputation models can be specified for different variables. This is useful if there are particular properties of the data that need to be preserved. However, the flexibility of the approach can also cause estimation problems. Specifying different distributions can lead to slow convergence or non-convergence of the imputation model (See the “Compatability of Conditionals” and “Convergence of MICE” sections in the Stata help file on mi impute chained). Additionally, issues of complete and quasi-complete separation can happen when attempting to impute a large number of categorical variables. Overall, when attempting multiple imputation especially with MICE you should allow yourself sufficient time to build an appropriate model and time for modifications should convergence and/or estimation problems occur with your imputation model. The goal is to only have to go through this process once!
Other issues
1. Why Auxiliary variables?
So one question you may be asking yourself, is why are auxiliary variables necessary or even important. First, they can help improve the likelihood of meeting the MAR assumption (White et al, 2011; Johnson and Young, 2011; Allison, 2012). Remember, a variable is said to be missing at random if other variables in the dataset can be used to predict missingness on a given variable. So you want your imputation model to include all the variables you think are associated with or predict missingness in your variable in order to fulfill the assumption of MAR. Second, including auxiliaries has been shown to help yield more accurate and stable estimates and thus reduce the estimated standard errors in analytic models (Enders, 2010; Allison, 2012; von Hippel and Lynch, 2013). This is especially true in the case of missing outcome variables. Third, including these variable can also help to increase power (Reis and Judd, 2000; Enders, 2010). In general, there is almost always a benefit to adopting a more “inclusive analysis strategy” (Enders, 2010; Allison, 2012).
2. Selecting the number of imputations (m) Historically, the recommendation was for three to five MI datasets. Relatively low values of m may still be appropriate when the fraction of missing information is low and the analysis techniques are relatively simple. Recently, however, larger values of m are often being recommended. To some extent, this change in the recommended number of imputations is based on the radical increase in the computing power available to the typical researcher, making it more practical to run, create and analyze multiply imputed datasets with a larger number of imputations. Recommendations for the number of m vary. For example, five to twenty imputations for low fractions of missing information, and as many as 50 (or more) imputations when the proportion of missing data is relatively high. Remember that estimates of coefficients stabilize at much lower values of m than estimates of variances and covariances of error terms (i.e., standard errors). Thus, in order to get appropriate estimates of these parameters, you may need to increase the m. A larger number of imputations may also allow hypothesis tests with less restrictive assumptions (i.e., that do not assume equal fractions of missing information for all coefficients). Multiple runs of m imputations are recommended to assess the stability of the parameter estimates.
Graham et al., 2007 conducted a simulation demonstrating the affect on power, efficiency and parameter estimates across different fractions of missing information as you decrease m. The authors found that:
1. Mean square error and standard error increased.
2. Power was reduced, especially when FMI is greater than 50% and the effect size is small, even for a large number of m (20 or more).
3. Variability of the estimate of FMI increased substantially. In general , the estimation of FMI improves with an increased m.
4. RE decreased.
Another factor to consider is the importance of reproducibility between analyses using the same data. White et al. (2010), assuming the true FMI for any variable would be less than or equal to the percentage of cases that are incomplete, uses the rule that m should equal the percentage of incomplete cases. Thus if the FMI for a variable is 20% then you need 20 imputed datasets. A similar analysis by Bodner, 2008 makes a similar recommendation. White et al., 2010 also found when making this assumption, the error associated with estimating the regression coefficients, standard errors and the resulting p-values was considerably reduced and resulted in an adequate level of reproducibility.
3. Maximum, Minimum and Round
This issue often comes up in the context of using MVN to impute variables that normally have integer values or bounds. Intuitively speaking, it makes sense to round values or incorporate bounds to give “plausible” values. However, these methods has been shown to decrease efficiency and increase bias by altering the correlation or covariances between variables estimated during the imputation process. Additionally, these changes will often result in an underestimation of the uncertainty around imputed values. Remember imputed values are NOT equivalent to observed values and serve only to help estimate the covariances between variables needed for inference (Johnson and Young 2011).
Leaving the imputed values as is in the imputation model is perfectly fine for your analytic models. If plausible values are needed to perform a specific type of analysis, then you may want to use a different imputation algorithm such as MICE.
4. Common questions?
Isn’t multiple imputation just making up data?
No. This is argument can be made of the missing data methods that use a single imputed value because this value will be treated like observed data, but this is not true of multiple imputation. Unlike single imputation, multiple imputation builds into the model the uncertainty/error associated with the missing data. Therefore the process and subsequent estimation never depends on a single value. Additionally, another method for dealing the missing data, maximum likelihood produces very similar results to multiple imputation and it does not require the missing information to be filled-in.
What is Passive imputation?
Passive variables are functions of imputed variables. For example, let’s say we have a variable X with missing information but in my analytic model we will need to use X2. In passive imputation we would impute X and then use those imputed values to create a quadratic term. This method is called “impute then transform” (von Hippel, 2009). While this appears to make sense, additional research (Seaman et al., 2012; Bartlett et al., 2014) has shown that using this method is actually a misspecification of your imputation model and will lead to biased parameter estimates in your analytic model. There are better ways of dealing with transformations.
How do I treat variable transformations such as logs, quadratics and interactions?
Most of the current literature on multiple imputation supports the method of treating variable transformations as “just another variable”. For example, if you know that in your subsequent analytic model you are interesting in looking at the modifying effect of Z on the association between X and Y (i.e. an interaction between X and Z). This is a property of your data that you want to be maintained in the resulting imputed values Using something like passive imputation, where the interaction is created after you impute X and/or Z means that the filled-in values are imputed under a model assuming that Z is not a moderator of the association between X an Y. Thus, your imputation model is now misspecified and the effect modification (e.g. interaction) of interest will be attenuated.
Should I include my dependent variable (DV) in my imputation model?
Yes! An emphatic YES unless you would like to impute independent variables (IVs) assuming they are uncorrelated with your DV (Enders, 2010). Thus, causing the estimated association between you DV and IV’s to be biased toward the null (i.e. underestimated).
Additionally, using imputed values of your DV is considered perfectly acceptable when you have good auxiliary variables in your imputation model (Enders, 2010; Johnson and Young, 2011; White et al., 2010). However, if good auxiliary variables are not available then you still INCLUDE your DV in the imputation model and then later restrict your analysis to only those observations with an observed DV value. Research has shown that imputing DV’s when auxiliary variables are not present can add unnecessary random variation into your imputed values (Allison, 2012) .
How much missing can I have and still get good estimates using MI?
Simulations have indicated that MI can perform well, under certain circumstances, even up to 50% missing observations (Allison, 2002). However, the larger the amount of missing information the higher the chance you will run into estimation problems during the imputation process and the lower the chance of meeting the MAR assumption unless it was planned missing (Johnson and Young, 2011). Additionally, as discussed further, the higher the FMI the more imputations are needed to reach good relative efficiency for effect estimates, especially standard errors.
What should I report in my methods abut my imputation?
Most papers mention if they performed multiple imputation but give very few if any details of how they implemented the method. In general, a basic description should include:
- Which statistical program was used to conduct the imputation.
- The type of imputation algorithm used (i.e. MVN or MICE).
- Some justification for choosing a particular imputation method.
- The number of imputed datasets (m) created.
- The proportion of missing observations for each imputed variable.
- The variables used in the imputation model and why so your audience will know if you used a more inclusive strategy. This is particularly important when using auxiliary variables.
This may seem like a lot, but probably would not require more than 4-5 sentences. Enders (2010) provides some examples of write-ups for particular scenarios. Additionally, MacKinnon (2010) discusses how to report MI procedures in medical journals.
Main Take Aways from this seminar:
- Multiple Imputation is always superior to any of the single imputation
methods because:
- A single imputed value is never used
- The variance estimates reflect the appropriate amount of uncertainty surrounding parameter estimates
- There are several decisions to be made before performing a multiple imputation including choice of distribution, auxiliary variables and number of imputations that can affect the quality of the imputation.
- Remember that multiple imputation is not magic, and while it can help increase power it should not be expected to provide “significant” effects when other techniques like listwise deletion fail to find significant associations.
- Multiple Imputation is one tool for researchers to address the very common problem of missing data.
References:
- Allison (2002). Missing Data. Sage Publications.
- Allison (2012). Handling Missing Data by Maximum Likelihood. SAS Global Forum: Statistics and Data Analysis.
- Allison (2005). Imputation of Categorical Variables with PROC MI. SUGI 30 Proceedings – Philadelphia, Pennsylvania April 10-13, 2005.
- Barnard and Rubin (1999). Small-sample degrees of freedom with multiple imputation. Biometrika, 86(4), 948-955.
- Bartlett et al. (2014). Multiple imputation of covariates by fully conditional specification: Accommodating the substantive model. Stat Methods Med Res.
- Bodner, T.E. (2008).”What Improves with Increased Missing Data Imputations?”. Structural Equation Modeling: A Multidisciplinary Journal, 15:4, 651-675.
- Demirtas et al.(2008). Plausibility of multivariate normality assumption when multiply imputing non-Gaussian continuous outcomes: a simulation assessment. Jour of Stat Computation & Simulation, 78(1).
- Enders (2010). Applied Missing Data Analysis. The Guilford Press.
- Graham et al. (2007). How Many Imputations are Really Needed? Some Practical Clarifications of Multiple Imputation Theory. Prev Sci, 8: 206-213.
- Horton et al. (2003) A potential for bias when rounding in multiple imputation. American Statistician. 57: 229-232.
- Lee and Carlin (2010). Multiple Imputation for missing data: Fully Conditional Specification versus Multivariate Normal Imputation. Am J Epidemiol, 171(5): 624-32.
- Lipsitz et al. (2002). A Degrees-of-Freedom Approximation in Multiple Imputation. J Statist Comput Simul, 72(4): 309-318.
- Little and Rubin (2002). Statistical Analysis with Missing Data, 2nd edition, New York: John Wiley.
- Mackinnon (2010). The use and reporting of multiple imputation in medical research – a review. J Intern Med, 268: 586–593.
- Johnson and Young (2011). Towards Best Practices in analyzing Datasets with Missing Data: Comparisons and Recommendations. Journal of Marriage and Family, 73(5): 926-45.
- Editors: Harry T. Reis, Charles M. Judd (2000). Handbook of Research Methods in Social and Personality Psychology.
-
Rubin (1976). Inference and Missing Data. Biometrika 63 (3), 581-592.
- Rubin (1987). Multiple Imputation for Nonresponse in Surveys. J. Wiley & Sons, New York.
- Seaman et al. (2012). Multiple Imputation of missing covariates with non-linear effects: an evaluation of statistical methods. BMC Medical Research Methodology, 12(46).
- Schafer and Graham (2002) Missing data: our view of the state of the art. Psychol Methods, 7(2):147-77
- StataCorp. 2017. Stata 15 Base Reference Manual. College Station, TX: Stata Press.
- van Buuren (2007). Multiple imputation of discrete and continuous data by fully conditional specification. Statistical Methods in Medical Research, 16: 219–242 .
- von Hippel (2009). How to impute interactions, squares and other transformed variables. Sociol Methodol, 39:265-291.
- von Hippel and Lynch (2013). Efficiency Gains from Using Auxiliary Variables in Imputation. Cornell University Library.
- von Hippel (2013). Should a Normal Imputation Model be modified to Impute Skewed Variables. Sociological Methods & Research, 42(1):105-138.
- White et al. (2011). Multiple imputation using chained equations: Issues and guidance for practice. Statistics in Medicine, 30(4): 377-399.
- Young and Johnson (2011). Imputing the Missing Y’s: Implications for Survey Producers and Survey Users. Proceedings of the AAPOR Conference Abstracts, pp. 6242–6248.